5DJU
 
 | | Crystal structure of LOV2 (C450A) domain in complex with Zdk3 | | Descriptor: | CHLORIDE ION, Engineered protein, Zdk3 affibody, ... | | Authors: | Tarnawski, M, Wang, H, Yumerefendi, H, Hahn, K.M, Schlichting, I. | | Deposit date: | 2015-09-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | LOVTRAP: an optogenetic system for photoinduced protein dissociation.
Nat.Methods, 13, 2016
|
|
5DJT
 
 | | Crystal structure of LOV2 (C450A) domain in complex with Zdk2 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Engineered protein, ... | | Authors: | Tarnawski, M, Wang, H, Yumerefendi, H, Hahn, K.M, Schlichting, I. | | Deposit date: | 2015-09-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | LOVTRAP: an optogenetic system for photoinduced protein dissociation.
Nat.Methods, 13, 2016
|
|
4I15
 
 | | Crystal structure of TbrPDEB1 | | Descriptor: | Class 1 phosphodiesterase PDEB1, MAGNESIUM ION, ZINC ION | | Authors: | Wang, H, Ke, H. | | Deposit date: | 2012-11-20 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of Novel Trypanosoma brucei Phosphodiesterase B1 Inhibitors by Virtual Screening against the Unliganded TbrPDEB1 Crystal Structure.
J.Med.Chem., 56, 2013
|
|
4IQ8
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 3 from Saccharomyces cerevisiae | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase 3 | | Authors: | Wang, H, Liu, Q, Niu, L, Teng, M, Li, X. | | Deposit date: | 2013-01-11 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Preliminary crystallographic analysis of glyceraldehyde-3-phosphate dehydrogenase 3 from Saccharomyces cerevisiae.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
8JHO
 
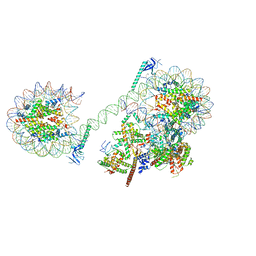 | |
5EFW
 
 | | Crystal structure of LOV2-Zdk1 - the complex of oat LOV2 and the affibody protein Zdark1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, NPH1-1, SULFATE ION, ... | | Authors: | Winkler, A, Wang, H, Hartmann, E, Hahn, K, Schlichting, I. | | Deposit date: | 2015-10-26 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | LOVTRAP: an optogenetic system for photoinduced protein dissociation.
Nat.Methods, 13, 2016
|
|
2DUC
 
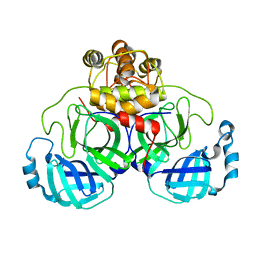 | | Crystal structure of SARS coronavirus main proteinase(3CLPRO) | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Wang, H, Kim, Y.T, Muramatsu, T, Takemoto, C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-21 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5WH5
 
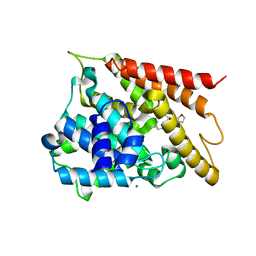 | |
5B6O
 
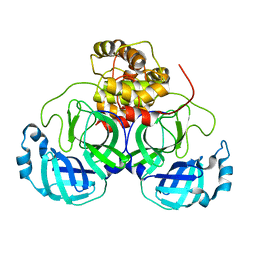 | | Crystal structure of MS8104 | | Descriptor: | 3C-like proteinase | | Authors: | Wang, H, Kim, Y, Muramatsu, T, Takemoto, C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-05-31 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
4Q4C
 
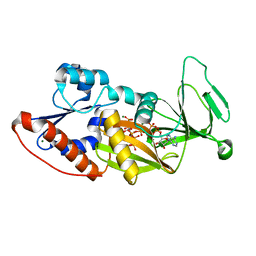 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP and synthetic 1,5-(PP)2-IP4 (1,5-IP8) | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], ADENOSINE-5'-DIPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase 2, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2014-04-14 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis of Densely Phosphorylated Bis-1,5-Diphospho-myo-Inositol Tetrakisphosphate and its Enantiomer by Bidirectional P-Anhydride Formation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4Q4D
 
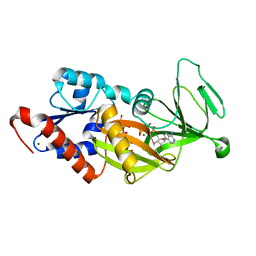 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMP-PNP and synthetic 3,5-(PP)2-IP4 (3,5-IP8) | | Descriptor: | (1R,3S,4S,5R,6S)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase 2, MAGNESIUM ION, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2014-04-14 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis of Densely Phosphorylated Bis-1,5-Diphospho-myo-Inositol Tetrakisphosphate and its Enantiomer by Bidirectional P-Anhydride Formation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5BYA
 
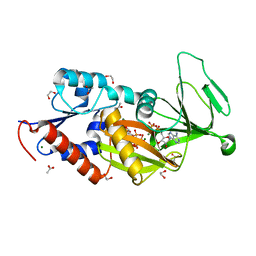 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP and 1,5-(PCP)2-IP4 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2015-06-10 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthetic tools for studying the chemical biology of InsP8.
Chem.Commun.(Camb.), 51, 2015
|
|
5BYB
 
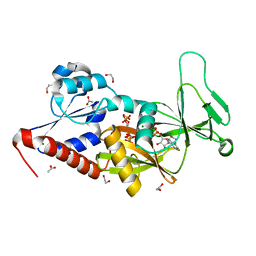 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP and 1,5-(PA)2-IP4 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2015-06-10 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthetic tools for studying the chemical biology of InsP8.
Chem.Commun.(Camb.), 51, 2015
|
|
5DGH
 
 | |
5DGI
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP and 3,5-(PCP)2-IP4 | | Descriptor: | 1,2-ETHANEDIOL, 3,5-di-methylenebisphosphonate inositol tetrakisphosphate, ACETATE ION, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2015-08-27 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cellular Cations Control Conformational Switching of Inositol Pyrophosphate Analogues.
Chemistry, 22, 2016
|
|
6LHE
 
 | | Crystal Structure of Gold-bound NDM-1 | | Descriptor: | GOLD ION, Metallo-beta-lactamase type 2, SULFATE ION | | Authors: | Wang, H, Sun, H, Wang, M. | | Deposit date: | 2019-12-07 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.206 Å) | | Cite: | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
8H3G
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166V Mutant in Complex with Inhibitor Enstrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, GLYCEROL | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3L
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (T21I and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3K
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, ... | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
5F7H
 
 | | Human T-cell immunoglobulin and mucin domain protein 4 (hTIM-4) complex with phosphoserine | | Descriptor: | CALCIUM ION, PHOSPHOSERINE, T-cell immunoglobulin and mucin domain-containing protein 4 | | Authors: | Gao, G.F, Lu, G, Wang, H, Qi, J. | | Deposit date: | 2015-12-08 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human TIM members: Ebolavirus entry-enhancing receptors
Chin.Sci.Bull., 60, 2015
|
|
4HN2
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMPPNP and a substrate analog 5PA-IP5 | | Descriptor: | (2-oxo-2-{[(1s,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl]oxy}ethyl)phosphonic acid, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase 2, MAGNESIUM ION, ... | | Authors: | Wang, H. | | Deposit date: | 2012-10-18 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First synthetic analogues of diphosphoinositol polyphosphates: interaction with PP-InsP(5) kinase.
Chem.Commun.(Camb.), 48, 2012
|
|
6AQ4
 
 | | CRYSTAL STRUCTURE OF PROTEIN CiTE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MAGNESIUM, PYRUVATE AND CITRAMALYL-COA | | Descriptor: | CHLORIDE ION, Citramalyl-CoA, Citrate lyase subunit beta-like protein, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wang, H, Bonanno, J.B, Carvalho, L, Almo, S.C. | | Deposit date: | 2017-08-18 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.825 Å) | | Cite: | An essential bifunctional enzyme inMycobacterium tuberculosisfor itaconate dissimilation and leucine catabolism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6AS5
 
 | | CRYSTAL STRUCTURE OF PROTEIN CitE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MAGNESIUM, ACETOACETATE AND COENZYME A | | Descriptor: | ACETOACETIC ACID, COENZYME A, Citrate lyase subunit beta-like protein, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wang, H, Bonanno, J.B, Carvalho, L, Almo, S.C. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.035 Å) | | Cite: | An essential bifunctional enzyme inMycobacterium tuberculosisfor itaconate dissimilation and leucine catabolism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6ARB
 
 | | CRYSTAL STRUCTURE OF PROTEIN CitE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MAGNESIUM, PYRUVATE AND COENZYME A | | Descriptor: | COENZYME A, Citrate lyase subunit beta-like protein, GLYCEROL, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wang, H, Bonanno, J.B, Carvalho, L, Almo, S.C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | An essential bifunctional enzyme inMycobacterium tuberculosisfor itaconate dissimilation and leucine catabolism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5F7F
 
 | |
