3ARN
 
 | | Human dUTPase in complex with novel uracil derivative | | Descriptor: | Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION, N-{5-[(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)methoxy]-2-methylpentan-2-yl}benzenesulfonamide | | Authors: | Chong, K.T, Miyahara, S, Miyakoshi, H, Fukuoka, M. | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a novel class of potent human deoxyuridine triphosphatase inhibitors remarkably enhancing the antitumor activity of thymidylate synthase inhibitors
J.Med.Chem., 55, 2012
|
|
1JXF
 
 | | SOLUTION STRUCTURE OF REDUCED CU(I) PLASTOCYANIN FROM SYNECHOCYSTIS PCC6803 | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bertini, I, Bryant, D.A, Ciurli, S, Dikiy, A, Fernandez, C.O, Luchinat, C, Safarov, N, Vila, A.J, Zhao, J. | | Deposit date: | 2001-09-07 | | Release date: | 2001-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics of plastocyanin in both oxidation states. Solution structure of the reduced form and comparison with the oxidized state.
J.Biol.Chem., 276, 2001
|
|
1JXD
 
 | | SOLUTION STRUCTURE OF REDUCED CU(I) PLASTOCYANIN FROM SYNECHOCYSTIS PCC6803 | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bertini, I, Bryant, D.A, Ciurli, S, Dikiy, A, Fernandez, C.O, Luchinat, C, Safarov, N, Vila, A.J, Zhao, J. | | Deposit date: | 2001-09-07 | | Release date: | 2001-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics of plastocyanin in both oxidation states. Solution structure of the reduced form and comparison with the oxidized state.
J.Biol.Chem., 276, 2001
|
|
1CKO
 
 | |
3ARA
 
 | | Discovery of Novel Uracil Derivatives as Potent Human dUTPase Inhibitors | | Descriptor: | 1-[3-({(2R)-2-[hydroxy(diphenyl)methyl]pyrrolidin-1-yl}sulfonyl)propyl]pyrimidine-2,4(1H,3H)-dione, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Chong, K.T, Miyakoshi, H, Miyahara, S, Fukuoka, M. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and discovery of N-carbonylpyrrolidine- or N-sulfonylpyrrolidine-containing uracil derivatives as potent human deoxyuridine triphosphatase inhibitors
J.Med.Chem., 55, 2012
|
|
2NA1
 
 | | ULD complex | | Descriptor: | Polycomb complex protein BMI-1, Polyhomeotic-like 2 | | Authors: | Cierpicki, T, Gray, F, Cho, H. | | Deposit date: | 2015-12-17 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | BMI1 regulates PRC1 architecture and activity through homo- and hetero-oligomerization.
Nat Commun, 7, 2016
|
|
2Q8X
 
 | | The high-resolution crystal structure of ixt6, a thermophilic, intracellular xylanase from G. stearothermophilus | | Descriptor: | GLYCEROL, SODIUM ION, intra-cellular xylanase | | Authors: | Solomon, V, Teplitsky, A, Gilboa, R, Zolotnitsky, G, Golan, G, Reiland, V, Moryles, S, Shoham, Y, Shoham, G. | | Deposit date: | 2007-06-12 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-specificity relationships of an intracellular xylanase from Geobacillus stearothermophilus
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3EF0
 
 | |
3EF1
 
 | |
3MHA
 
 | | Crystal structure of LprG from Mycobacterium tuberculosis bound to PIM | | Descriptor: | (1S,2R,3R,4S,5R,6S)-2-(alpha-L-allopyranosyloxy)-3,4,5-trihydroxy-6-({6-O-[(1R)-1-hydroxyhexadecyl]-beta-L-gulopyranosyl}oxy)cyclohexyl (2R)-2-{[(1S)-1-hydroxyhexadecyl]oxy}-3-{[(1S,10S)-1-hydroxy-10-methyloctadecyl]oxy}propyl hydrogen (R)-phosphate, Lipoprotein lprG | | Authors: | Tsai, H.-C, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-04-07 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mycobacterium tuberculosis lipoprotein LprG (Rv1411c) binds triacylated glycolipid agonists of Toll-like receptor 2.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MH8
 
 | |
7JTW
 
 | | Crystal structure of RORgt with compound (4R)-6-[(2,5-dichloro-3-{[(2R,4R)-1-(cyclopentanecarbonyl)-2-methylpiperidin-4-yl]oxy}phenyl)amino]-6-oxo-4-phenylhexanoic acid | | Descriptor: | (4R)-6-[(2,5-dichloro-3-{[(2R,4R)-1-(cyclopentanecarbonyl)-2-methylpiperidin-4-yl]oxy}phenyl)amino]-6-oxo-4-phenylhexanoic acid, GLYCEROL, RAR-related orphan receptor C isoform a variant, ... | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2020-08-18 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of 6-Oxo-4-phenyl-hexanoic acid derivatives as ROR gamma t inverse agonists showing favorable ADME profile.
Bioorg.Med.Chem.Lett., 36, 2021
|
|
2HVQ
 
 | | Structure of Adenylated full-length T4 RNA Ligase 2 | | Descriptor: | Hypothetical 37.6 kDa protein in Gp24-hoc intergenic region, MAGNESIUM ION | | Authors: | Nandakumar, J, Lima, C.D. | | Deposit date: | 2006-07-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
2HVS
 
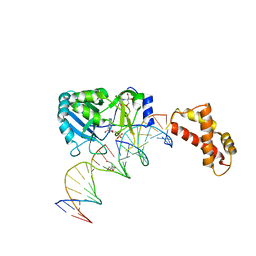 | | Structure of T4 RNA Ligase 2 with Nicked 5'-Adenylated nucleic acid duplex containing a 2'-deoxyribonucleotide at the nick | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*TP*TP*G)-3', 5'-D(*CP*AP*AP*TP*TP*GP*CP*GP*AP*C)-R(P*(OMC)P*C)-3', ... | | Authors: | Nandakumar, J, Lima, C.D. | | Deposit date: | 2006-07-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
2HVR
 
 | | Structure of T4 RNA Ligase 2 with Nicked 5'-Adenylated nucleic acid duplex containing a 3'-deoxyribonucleotide at the nick | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*TP*TP*G)-3', 5'-D(*CP*AP*AP*TP*TP*GP*CP*GP*AP*C)-R(P*(OMC)P*C)-3', ... | | Authors: | Nandakumar, J, Lima, C.D. | | Deposit date: | 2006-07-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
4LOH
 
 | | Crystal structure of hSTING(H232) in complex with c[G(2',5')pA(3',5')p] | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA.
Cell(Cambridge,Mass.), 154, 2013
|
|
4LOJ
 
 | | Crystal structure of mSting in complex with c[G(2',5')pA(3',5')p] | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA.
Cell(Cambridge,Mass.), 154, 2013
|
|
4LOL
 
 | | Crystal structure of mSting in complex with DMXAA | | Descriptor: | (5,6-dimethyl-9-oxo-9H-xanthen-4-yl)acetic acid, Stimulator of interferon genes protein | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA.
Cell(Cambridge,Mass.), 154, 2013
|
|
4LOI
 
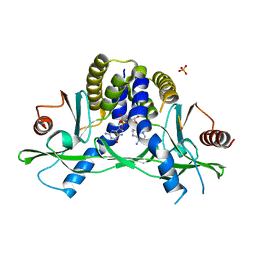 | | Crystal structure of hSTING(H232) in complex with c[G(2',5')pA(2',5')p] | | Descriptor: | 2-amino-9-[(1R,3R,6R,8R,9R,11S,14R,16R,17R,18R)-16-(6-amino-9H-purin-9-yl)-3,11,17,18-tetrahydroxy-3,11-dioxido-2,4,7,10,12,15-hexaoxa-3,11-diphosphatricyclo[12.2.1.1~6,9~]octadec-8-yl]-1,9-dihydro-6H-purin-6-one, PHOSPHATE ION, Stimulator of interferon genes protein | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA.
Cell(Cambridge,Mass.), 154, 2013
|
|
4LOK
 
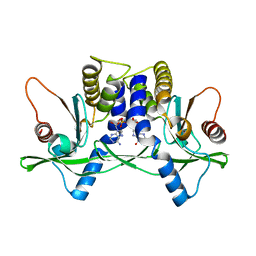 | | Crystal structure of mSting in complex with c[G(3',5')pA(3',5')p] | | Descriptor: | 2-amino-9-[(2R,3R,3aR,5S,7aS,9R,10R,10aR,12R,14aS)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Stimulator of interferon genes protein | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA.
Cell(Cambridge,Mass.), 154, 2013
|
|
3RTX
 
 | |
3GAT
 
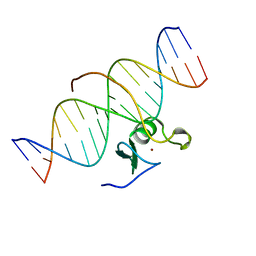 | | SOLUTION NMR STRUCTURE OF THE C-TERMINAL DOMAIN OF CHICKEN GATA-1 BOUND TO DNA, 34 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*TP*TP*AP*TP*CP*TP*GP*CP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*AP*GP*AP*TP*AP*AP*AP*CP*AP*TP*T)-3'), ERYTHROID TRANSCRIPTION FACTOR GATA-1, ... | | Authors: | Clore, G.M, Tjandra, N, Starich, M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution.
Nat.Struct.Biol., 4, 1997
|
|
3U4I
 
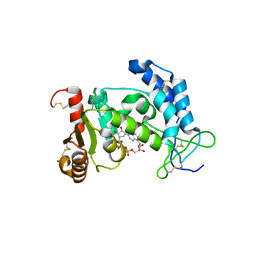 | | CD38 structure-based inhibitor design using the N1-cyclic inosine 5'-diphosphate ribose template | | Descriptor: | ADP-ribosyl cyclase 1, Cyclic adenosine 5'-diphosphocarbocyclic ribose | | Authors: | Liu, Q, Hao, Q, Lee, H.C, Graeff, R. | | Deposit date: | 2011-10-08 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.118 Å) | | Cite: | CD38 Structure-Based Inhibitor Design Using the N1-Cyclic Inosine 5'-Diphosphate Ribose Template
Plos One, 8, 2013
|
|
3U4H
 
 | | CD38 structure-based inhibitor design using the N1-cyclic inosine 5'-diphosphate ribose template | | Descriptor: | 8-Amino-N1-Cyclic Inosine 5'-Diphosphoribose, ADP-ribosyl cyclase 1 | | Authors: | Liu, Q, Hao, Q, Lee, H.C, Graeff, R. | | Deposit date: | 2011-10-08 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | CD38 Structure-Based Inhibitor Design Using the N1-Cyclic Inosine 5'-Diphosphate Ribose Template
Plos One, 8, 2013
|
|
