5H5N
 
 | |
5GSV
 
 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 142-5 | | Descriptor: | 10-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GR7
 
 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 37-1 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-D alpha chain, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-08 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GSR
 
 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide I5A | | Descriptor: | 9-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GYQ
 
 | |
5GSB
 
 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 37-3 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-D alpha chain, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GZN
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody Heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
5HGB
 
 | | HLA*A2402 complexed with HIV nef138 8mer epitope | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Effects of a Single Escape Mutation on T Cell and HIV-1 Co-adaptation.
Cell Rep, 15, 2016
|
|
5HGD
 
 | | HLA*A2402 complexed with HIV nef138 Y2F mutant 10mer epitope | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Effects of a Single Escape Mutation on T Cell and HIV-1 Co-adaptation.
Cell Rep, 15, 2016
|
|
5HGA
 
 | | HLA*A2402 complex with HIV nef138 Y2F-8mer mutant epitope | | Descriptor: | 8-mer from Protein Nef, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Effects of a Single Escape Mutation on T Cell and HIV-1 Co-adaptation.
Cell Rep, 15, 2016
|
|
7YE7
 
 | | Crystal structure of SARS-CoV-2 soluble dimeric ORF9b | | Descriptor: | N-OCTANE, ORF9b protein, nonane | | Authors: | Jin, X, Chai, Y, Qi, J, Song, H, Gao, G.F. | | Deposit date: | 2022-07-05 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural characterization of SARS-CoV-2 dimeric ORF9b reveals potential fold-switching trigger mechanism.
Sci China Life Sci, 66, 2023
|
|
5IY3
 
 | | Zika Virus Non-structural Protein NS1 | | Descriptor: | Genome polyprotein | | Authors: | Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2016-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Zika virus NS1 structure reveals diversity of electrostatic surfaces among flaviviruses
Nat.Struct.Mol.Biol., 23, 2016
|
|
8WP8
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 RBD in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-10-09 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 RBD in complex with human ACE2
To Be Published
|
|
8WOY
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 RBD in complex with rabbit ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Li, L.J, Shi, K.Y, Yu, G.H, Gao, G.F. | | Deposit date: | 2023-10-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis of increased binding affinities of spikes from SARS-CoV-2 Omicron variants to rabbit and hare ACE2s reveals the expanding host tendency.
Mbio, 15, 2024
|
|
8WOZ
 
 | | Cryo-EM structure of SARS-CoV RBD in complex with rabbit ACE2 | | Descriptor: | Angiotensin-converting enzyme, Spike protein S1, ZINC ION | | Authors: | Li, L.J, Shi, K.Y, Yu, G.H, Gao, G.F. | | Deposit date: | 2023-10-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis of increased binding affinities of spikes from SARS-CoV-2 Omicron variants to rabbit and hare ACE2s reveals the expanding host tendency.
Mbio, 15, 2024
|
|
8WOX
 
 | | Cryo-EM structure of SARS-CoV-2 prototype RBD in complex with rabbit ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Li, L.J, Shi, K.Y, Yu, G.H, Gao, G.F. | | Deposit date: | 2023-10-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural basis of increased binding affinities of spikes from SARS-CoV-2 Omicron variants to rabbit and hare ACE2s reveals the expanding host tendency.
Mbio, 15, 2024
|
|
5HGH
 
 | | HLA*A2402 complexed with HIV nef138 10mer epitope | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Effects of a Single Escape Mutation on T Cell and HIV-1 Co-adaptation.
Cell Rep, 15, 2016
|
|
8XB8
 
 | | The structure of ASFV A137R | | Descriptor: | A137R | | Authors: | Li, C, Song, H, Gao, G.F. | | Deposit date: | 2023-12-06 | | Release date: | 2024-01-31 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | African swine fever virus A137R assembles into a dodecahedron cage.
J.Virol., 98, 2024
|
|
7YE8
 
 | | Crystal structure of SARS-CoV-2 refolded dimeric ORF9b | | Descriptor: | N-OCTANE, ORF9b protein | | Authors: | Jin, X, Chai, Y, Qi, J, Song, H, Gao, G.F. | | Deposit date: | 2022-07-05 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural characterization of SARS-CoV-2 dimeric ORF9b reveals potential fold-switching trigger mechanism.
Sci China Life Sci, 66, 2023
|
|
5JHL
 
 | | Crystal structure of zika virus envelope protein in complex with a flavivirus broadly-protective antibody | | Descriptor: | Antibody Heavy chain, antibody Light chain, envelope protein | | Authors: | Dai, L, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2016-04-21 | | Release date: | 2016-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the Zika Virus Envelope Protein and Its Complex with a Flavivirus Broadly Protective Antibody.
Cell Host Microbe, 19, 2016
|
|
8K4U
 
 | |
8KA8
 
 | | Cryo-EM structure of SARS-CoV-2 Delta RBD in complex with golden hamster ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Niu, S, Zhao, Z.N, Chai, Y, Gao, G.F. | | Deposit date: | 2023-08-02 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural basis and analysis of hamster ACE2 binding to different SARS-CoV-2 spike RBDs.
J.Virol., 98, 2024
|
|
8KC2
 
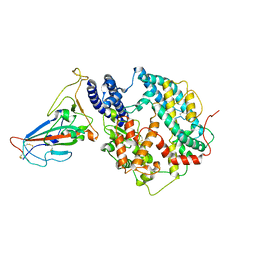 | | Cryo-EM structure of SARS-CoV-2 BA.3 RBD in complex with golden hamster ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Niu, S, Zhao, Z.N, Chai, Y, Gao, G.F. | | Deposit date: | 2023-08-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis and analysis of hamster ACE2 binding to different SARS-CoV-2 spike RBDs.
J.Virol., 98, 2024
|
|
5WZ1
 
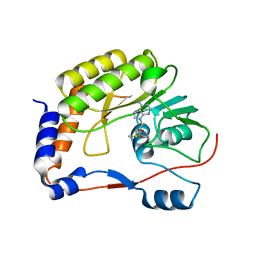 | | Crystal structure of Zika virus NS5 methyltransferase bound to S-adenosyl-L-methionine | | Descriptor: | NS5 methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Duan, W, Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | The crystal structure of Zika virus NS5 reveals conserved drug targets.
EMBO J., 36, 2017
|
|
5WWI
 
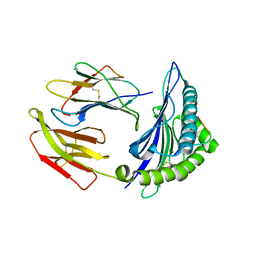 | | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 1) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2017-01-01 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 1).
To Be Published
|
|
