4HD9
 
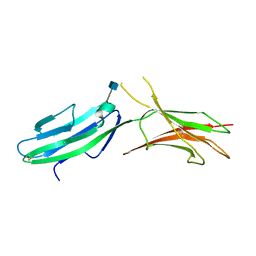 | | Crystal structure of native human MAdCAM-1 D1D2 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Mucosal addressin cell adhesion molecule 1 | | Authors: | Springer, T, Yu, Y, Zhu, J. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Different Fold with an Integrin-Binding Loop Specialized for Flexibility in Mucosal Addressin Cell Adhesion Molecule-1
TO BE PUBLISHED
|
|
4HC1
 
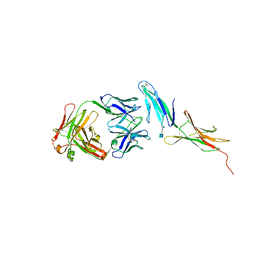 | |
4HBQ
 
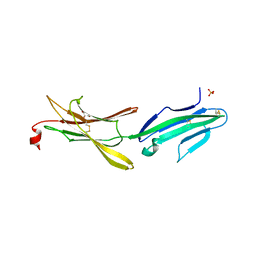 | | Crystal structure of a loop deleted mutant of Human MAdCAM-1 D1D2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Mucosal addressin cell adhesion molecule 1, ... | | Authors: | Springer, T, Yu, Y, Zhu, J. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-23 | | Last modified: | 2021-05-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Different Fold with an Integrin-Binding Loop Specialized for Flexibility in Mucosal Addressin Cell Adhesion Molecule-1
TO BE PUBLISHED
|
|
4JHR
 
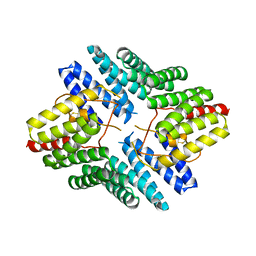 | | An auto-inhibited conformation of LGN reveals a distinct interaction mode between GoLoco motifs and TPR motifs | | Descriptor: | G-protein-signaling modulator 2 | | Authors: | Pan, Z, Zhu, J, Shang, Y, Wei, Z, Jia, M, Xia, C, Wen, W, Wang, W, Zhang, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An autoinhibited conformation of LGN reveals a distinct interaction mode between GoLoco motifs and TPR motifs
Structure, 21, 2013
|
|
6IF2
 
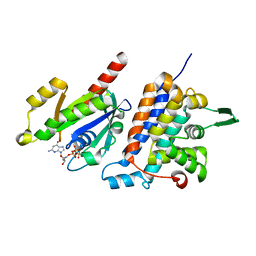 | | Complex structure of Rab35 and its effector RUSC2 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Iporin, MAGNESIUM ION, ... | | Authors: | Lin, L, Zhu, J, Zhang, R. | | Deposit date: | 2018-09-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rab35/ACAP2 and Rab35/RUSC2 Complex Structures Reveal Molecular Basis for Effector Recognition by Rab35 GTPase.
Structure, 27, 2019
|
|
6IF3
 
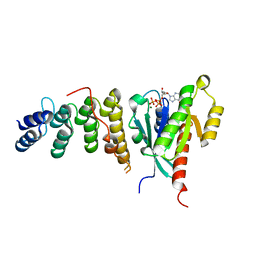 | | Complex structure of Rab35 and its effector ACAP2 | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lin, L, Zhu, J, Zhang, R. | | Deposit date: | 2018-09-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rab35/ACAP2 and Rab35/RUSC2 Complex Structures Reveal Molecular Basis for Effector Recognition by Rab35 GTPase.
Structure, 27, 2019
|
|
6JIQ
 
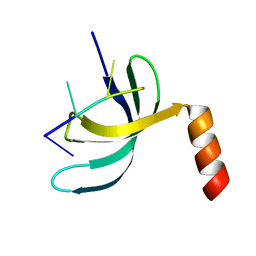 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT6 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*T)-3'), SP_0782 | | Authors: | Fang, X, Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
6JIP
 
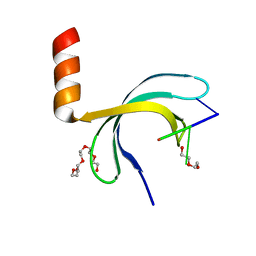 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT6 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*T)-3'), PENTAETHYLENE GLYCOL, SP_0782 | | Authors: | Fang, X, Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
6IYN
 
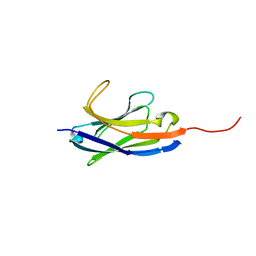 | | Solution structure of camelid nanobody Nb26 against aflatoxin B1 | | Descriptor: | Nb26 | | Authors: | Nie, Y, He, T, Zhu, J, Li, S.L, Hu, R, Yang, Y.H. | | Deposit date: | 2018-12-17 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Chemical shift assignments of a camelid nanobody against aflatoxin B1.
Biomol NMR Assign, 13, 2019
|
|
5XZR
 
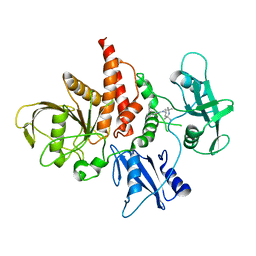 | | The atomic structure of SHP2 E76A mutant in complex with allosteric inhibitor 9b | | Descriptor: | 4-(3-phenylphenyl)-N-(2,2,6,6-tetramethylpiperidin-4-yl)-1,3-thiazol-2-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Li, D, Xie, J, Zhu, J, Liu, C. | | Deposit date: | 2017-07-13 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric Inhibitors of SHP2 with Therapeutic Potential for Cancer Treatment.
J. Med. Chem., 60, 2017
|
|
7WF6
 
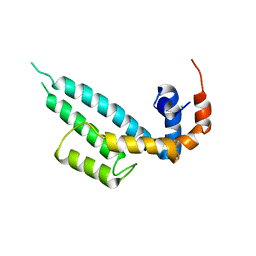 | | Crystal structure of SNX13 RGS domain | | Descriptor: | CHLORIDE ION, Sorting nexin-13 | | Authors: | Xu, J, Zhu, J, Liu, J. | | Deposit date: | 2021-12-26 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural Studies Reveal Unique Non-canonical Regulators of G Protein Signaling Homology (RH) Domains in Sorting Nexins.
J.Mol.Biol., 434, 2022
|
|
7U08
 
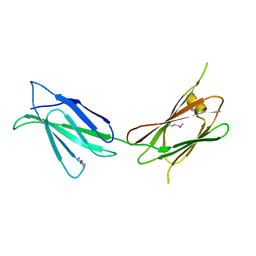 | |
7U01
 
 | |
6BXJ
 
 | | Structure of a single-chain beta3 integrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chimera protein of Integrin beta-3 and Integrin alpha-L, MAGNESIUM ION | | Authors: | Thinn, A.M.M, Wang, Z, Zhou, D, Zhao, Y, Curtis, B.R, Zhu, J. | | Deposit date: | 2017-12-18 | | Release date: | 2018-10-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Autonomous conformational regulation of beta3integrin and the conformation-dependent property of HPA-1a alloantibodies.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C45
 
 | |
6BXB
 
 | | Crystal structure of an extended b3 integrin P33 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Chimera protein of Integrin beta-3 and Integrin alpha-L, ... | | Authors: | Zhou, D, Zhu, J. | | Deposit date: | 2017-12-18 | | Release date: | 2018-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of an extended beta3integrin.
Blood, 132, 2018
|
|
6CKB
 
 | | Crystal structure of an extended beta3 integrin P33 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Chimera protein of Integrin beta-3 and Integrin alpha-L, ... | | Authors: | Zhou, D, Zhu, J. | | Deposit date: | 2018-02-27 | | Release date: | 2018-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an extended beta3integrin.
Blood, 132, 2018
|
|
6BXF
 
 | | Crystal structure of an extended b3 integrin L33 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Chimera protein of Integrin beta-3 and Integrin alpha-L, ... | | Authors: | Zhou, D, Zhu, J. | | Deposit date: | 2017-12-18 | | Release date: | 2018-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of an extended beta3integrin.
Blood, 132, 2018
|
|
6KZ1
 
 | | Complex structure of Whirlin and Myosin XVa | | Descriptor: | Myosin XVa, Whirlin | | Authors: | Lin, L, Wang, M, Shi, Y, Zhu, J, Zhang, R. | | Deposit date: | 2019-09-22 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Phase separation-mediated condensation of Whirlin-Myo15-Eps8 stereocilia tip complex.
Cell Rep, 34, 2021
|
|
7XHQ
 
 | | Small-molecule Allosteric Regulation Mechanism of SHP2 | | Descriptor: | 2-[(3S,4R)-4-azanyl-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-5-[2,3-bis(chloranyl)phenyl]-3-methyl-pyrrolo[2,1-f][1,2,4]triazin-4-one, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Luo, Y, Zhu, J, Yu, K, Liu, B. | | Deposit date: | 2022-04-09 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of the SHP2 allosteric inhibitor 2-((3R,4R)-4-amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl)-5-(2,3-dichlorophenyl)-3-methylpyrrolo[2,1-f][1,2,4] triazin-4(3H)-one.
J Enzyme Inhib Med Chem, 38, 2023
|
|
7EIL
 
 | | BRD4-BD1 in complex with LT-909-110 | | Descriptor: | Bromodomain-containing protein 4, N-[4-[4-ethanoyl-5-methyl-2-[6-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-pyrrol-3-yl]phenyl]ethanamide | | Authors: | Zheng, W, Kong, B, Tang, W, Zhu, J, Chen, Y. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of 1-(5-(1H-benzo[d]imidazole-2-yl)-2,4-dimethyl-1H-pyrrol-3-yl)ethan-1-one derivatives as novel and potent bromodomain and extra-terminal (BET) inhibitors with anticancer efficacy
Eur.J.Med.Chem., 227, 2022
|
|
8GYD
 
 | | Structure of Schistosoma japonicum Glutathione S-transferase bound with the ligand complex of 16 | | Descriptor: | (2R)-2-[[2-(5-chloranylthiophen-2-yl)-4-oxidanylidene-6-[2-(1H-1,2,3,4-tetrazol-5-yl)phenyl]quinazolin-3-yl]methyl]-3-(4-chlorophenyl)propanoic acid, ETHANOL, Glutathione S-transferase class-mu 26 kDa isozyme | | Authors: | Wen, X, Jin, R, Hu, H, Zhu, J, Song, W, Lu, X. | | Deposit date: | 2022-09-22 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery, SAR Study of GST Inhibitors from a Novel Quinazolin-4(1 H )-one Focused DNA-Encoded Library.
J.Med.Chem., 66, 2023
|
|
8GWW
 
 | | Small-molecule Allosteric Regulation Mechanism of SHP2 | | Descriptor: | 2-[4-(aminomethyl)-4-methyl-piperidin-1-yl]-5-[2,3-bis(chloranyl)phenyl]-3-methyl-pyrrolo[2,1-f][1,2,4]triazin-4-one, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Luo, Y, Zhu, J, Yu, K, Liu, B. | | Deposit date: | 2022-09-17 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Small-molecule Allosteric Regulation Mechanism of SHP2
To Be Published
|
|
7CFU
 
 | |
8HWD
 
 | | Cryo-EM Structure of D5 ADP form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Primase D5 | | Authors: | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | Deposit date: | 2022-12-29 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
