6FQB
 
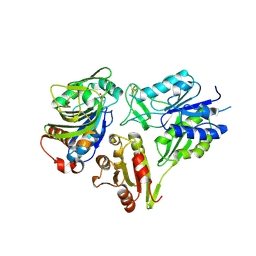 | | MurT/GatD peptidoglycan amidotransferase complex from Streptococcus pneumoniae R6 | | Descriptor: | Cobyric acid synthase, GLUTAMINE, Mur ligase family protein | | Authors: | Morlot, C, Contreras-Martel, C, Leisico, F, Straume, D, Peters, K, Hegnar, O.A, Simon, N, Villard, A.M, Breukink, E, Gravier-Pelletier, C, Le Corre, L, Vollmer, W, Pietrancosta, N, Havarstein, L.S, Zapun, A. | | Deposit date: | 2018-02-13 | | Release date: | 2018-08-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the essential peptidoglycan amidotransferase MurT/GatD complex from Streptococcus pneumoniae.
Nat Commun, 9, 2018
|
|
6XUC
 
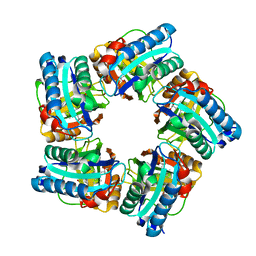 | | Structure of coproheme decarboxylase from Corynebacterium diphteriae in complex with coproheme | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Chlorite dismutase | | Authors: | Michlits, H, Lier, B, Pfanzagl, V, Djinovic-Carugo, K, Furtmueller, P.G, Oostenbrink, C, Obinger, C, Hofbauer, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8702 Å) | | Cite: | Actinobacterial Coproheme Decarboxylases Use Histidine as a Distal Base to Promote Compound I Formation.
Acs Catalysis, 10, 2020
|
|
8BAD
 
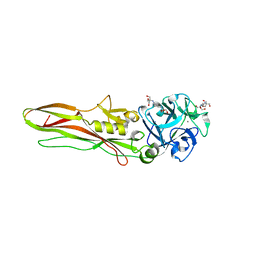 | | Tpp80Aa1 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Binary toxin A-like protein, CALCIUM ION, ... | | Authors: | Best, H.L, Williamson, L.J, Rizkallah, P.J, Berry, C, Lloyd-Evans, E. | | Deposit date: | 2022-10-11 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Crystal Structure of Bacillus thuringiensis Tpp80Aa1 and Its Interaction with Galactose-Containing Glycolipids.
Toxins, 14, 2022
|
|
8BBH
 
 | |
8BD5
 
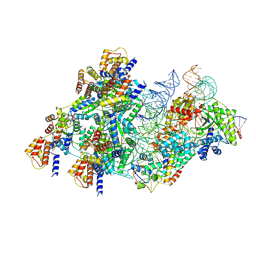 | | Cas12k-sgRNA-dsDNA-S15-TniQ-TnsC transposon recruitment complex | | Descriptor: | 30S ribosomal protein S15, ADENOSINE-5'-TRIPHOSPHATE, DNA non-target strand, ... | | Authors: | Schmitz, M, Querques, I, Oberli, S, Chanez, C, Jinek, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the assembly of the type V CRISPR-associated transposon complex.
Cell, 185, 2022
|
|
8B1U
 
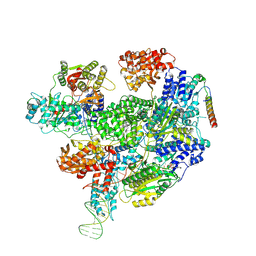 | | RecBCD-DNA in complex with the phage protein Abc2 and host PpiB | | Descriptor: | Anti-RecBCD protein 2, DNA (70-MER), MAGNESIUM ION, ... | | Authors: | Wilkinson, M, Wilkinson, O.J, Feyerherm, C, Fletcher, E.E, Wigley, D.B, Dillingham, M.S. | | Deposit date: | 2022-09-12 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of RecBCD in complex with phage-encoded inhibitor proteins reveal distinctive strategies for evasion of a bacterial immunity hub.
Elife, 11, 2022
|
|
8BLV
 
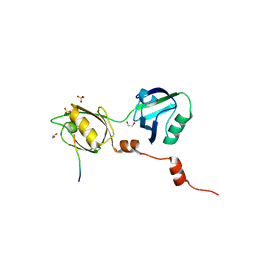 | | The PDZ domains of human SDCBP with a bound SDC4 C-terminal peptide | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Syndecan-4, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Daniel-Mozo, M, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2022-11-10 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The PDZ domains of human SDCBP with a bound SDC4 C-terminal peptide
To Be Published
|
|
8B1R
 
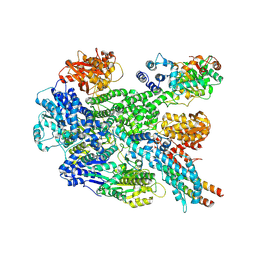 | | RecBCD in complex with the phage protein gp5.9 | | Descriptor: | MAGNESIUM ION, Probable RecBCD inhibitor gp5.9, RecBCD enzyme subunit RecB, ... | | Authors: | Wilkinson, M, Wilkinson, O.J, Feyerherm, C, Fletcher, E.E, Wigley, D.B, Dillingham, M.S. | | Deposit date: | 2022-09-12 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of RecBCD in complex with phage-encoded inhibitor proteins reveal distinctive strategies for evasion of a bacterial immunity hub.
Elife, 11, 2022
|
|
6XIW
 
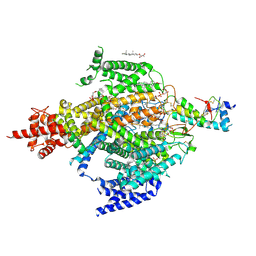 | | Cryo-EM structure of the sodium leak channel NALCN-FAM155A complex | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kschonsak, M, Chua, H.C, Noland, C.L, Weidling, C, Clairfeuille, T, Bahlke, O.O, Ameen, A.O, Li, Z.R, Arthur, C.P, Ciferri, C, Pless, S.A, Payandeh, J. | | Deposit date: | 2020-06-22 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human sodium leak channel NALCN.
Nature, 587, 2020
|
|
8BD4
 
 | | TniQ-capped Tns-ATP-dsDNA complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*GP*AP*TP*CP*GP*AP*TP*CP*GP*AP*TP*CP*GP*AP*TP*C)-3'), MAGNESIUM ION, ... | | Authors: | Querques, I, Schmitz, M, Oberli, S, Chanez, C, Jinek, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural basis for the assembly of the type V CRISPR-associated transposon complex.
Cell, 185, 2022
|
|
8BC6
 
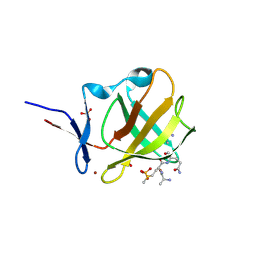 | |
8FW5
 
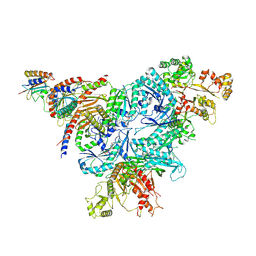 | | Chimeric HsGATOR1-SpGtr-SpLam complex | | Descriptor: | ALUMINUM FLUORIDE, GATOR complex protein DEPDC5, GATOR complex protein NPRL2, ... | | Authors: | Tettoni, S.D, Egri, S.B, Doxsey, D.D, Ouch, C, Chang, J, Song, K, Xu, C, Shen, K. | | Deposit date: | 2023-01-20 | | Release date: | 2023-07-26 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure of the Schizosaccharomyces pombe Gtr-Lam complex reveals evolutionary divergence of mTORC1-dependent amino acid sensing.
Structure, 31, 2023
|
|
8BC7
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex an aminoglutarimide degron peptide | | Descriptor: | Cereblon isoform 4, PHE-PHE-GLU-GLN-MET-GLN-QCI, S-Thalidomide, ... | | Authors: | Heim, C, Albrecht, R, Spring, A.K, Hartmann, M.D. | | Deposit date: | 2022-10-15 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Identification and structural basis of C-terminal cyclic imides as natural degrons for cereblon.
Biochem.Biophys.Res.Commun., 637, 2022
|
|
8B1T
 
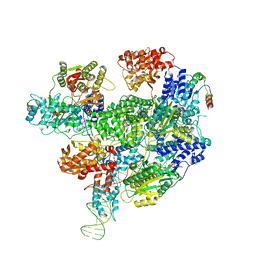 | | RecBCD-DNA in complex with the phage protein Abc2 | | Descriptor: | Anti-RecBCD protein 2, DNA (70-MER), MAGNESIUM ION, ... | | Authors: | Wilkinson, M, Wilkinson, O.J, Feyerherm, C, Fletcher, E.E, Wigley, D.B, Dillingham, M.S. | | Deposit date: | 2022-09-12 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of RecBCD in complex with phage-encoded inhibitor proteins reveal distinctive strategies for evasion of a bacterial immunity hub.
Elife, 11, 2022
|
|
8B8T
 
 | | Open conformation of the complex of DNA ligase I on PCNA and DNA in the presence of ATP | | Descriptor: | DNA ligase 1, Proliferating cell nuclear antigen | | Authors: | Blair, K, Tehseen, M, Raducanu, V.S, Shahid, T, Lancey, C, Cruehet, R, Hamdan, S, De Biasio, A. | | Deposit date: | 2022-10-05 | | Release date: | 2023-01-11 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanism of human Lig1 regulation by PCNA in Okazaki fragment sealing.
Nat Commun, 13, 2022
|
|
8ASG
 
 | | Structure of the SFTSV L protein bound in a resting state [RESTING] | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*AP*CP*AP*CP*AP*GP*AP*GP*AP*CP*GP*CP*CP*CP*AP*GP*AP*UP*GP*A*)-3'), RNA (5'-R(*GP*AP*UP*CP*UP*GP*GP*GP*CP*GP*GP*UP*CP*UP*UP*UP*GP*UP*GP*U*)-3'), ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Milewski, M, Busch, C, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2022-08-19 | | Release date: | 2023-01-18 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 51, 2023
|
|
7QLA
 
 | | Structure of the Rab GEF complex Mon1-Ccz1 | | Descriptor: | Ccz1, Vacuolar fusion protein MON1 | | Authors: | Klink, B.U, Herrmann, E, Antoni, C, Langemeyer, L, Kiontke, S, Gatsogiannis, C, Ungermann, C, Raunser, S, Kuemmel, D. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-09 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structure of the Mon1-Ccz1 complex reveals molecular basis of membrane binding for Rab7 activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8BG9
 
 | | Murine amyloid-beta filaments with the Arctic mutation (E22G) from APP(NL-G-F) mouse brains | ABeta | | Descriptor: | Amyloid-beta protein 40 | | Authors: | Yang, Y, Zhang, W.J, Murzin, A.G, Schweighauser, M, Huang, M, Lovestam, S.K.A, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Ghetti, B, Graff, C, Kumar, A, Nordber, A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2022-10-27 | | Release date: | 2023-01-18 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of amyloid-beta filaments with the Arctic mutation (E22G) from human and mouse brains.
Acta Neuropathol, 145, 2023
|
|
8BG0
 
 | | Amyloid-beta tetrameric filaments with the Arctic mutation (E22G) from Alzheimer's disease brains | ABeta40 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Yang, Y, Zhang, W.J, Murzin, A.G, Schweighauser, M, Huang, M, Lovestam, S.K.A, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Ghetti, B, Graff, C, Kumar, A, Nordber, A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2022-10-27 | | Release date: | 2023-01-18 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (1.99 Å) | | Cite: | Cryo-EM structures of amyloid-beta filaments with the Arctic mutation (E22G) from human and mouse brains.
Acta Neuropathol, 145, 2023
|
|
8BKD
 
 | | structure of RutB | | Descriptor: | N-(AMINOCARBONYL)-BETA-ALANINE, Ureidoacrylate amidohydrolase RutB | | Authors: | Rajendran, C. | | Deposit date: | 2022-11-09 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Functional Characterization of the Ureidoacrylate Amidohydrolase RutB from Escherichia coli .
Biochemistry, 62, 2023
|
|
8BFZ
 
 | | Amyloid-beta 42 filaments extracted from the human brain with Arctic mutation (E22G) of Alzheimer's disease | ABeta42 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Yang, Y, Zhang, W.J, Murzin, A.G, Schweighauser, M, Huang, M, Lovestam, S.K.A, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Ghetti, B, Graff, C, Kumar, A, Nordberg, A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2022-10-27 | | Release date: | 2023-01-18 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of amyloid-beta filaments with the Arctic mutation (E22G) from human and mouse brains.
Acta Neuropathol, 145, 2023
|
|
8BLM
 
 | | Structure of RutB | | Descriptor: | Ureidoacrylate amidohydrolase RutB | | Authors: | Rajendran, C. | | Deposit date: | 2022-11-09 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of the Ureidoacrylate Amidohydrolase RutB from Escherichia coli .
Biochemistry, 62, 2023
|
|
8C3X
 
 | |
8BLL
 
 | | Structure of RutB | | Descriptor: | ACETATE ION, Ureidoacrylate amidohydrolase RutB | | Authors: | Rajendran, C. | | Deposit date: | 2022-11-09 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural and Functional Characterization of the Ureidoacrylate Amidohydrolase RutB from Escherichia coli .
Biochemistry, 62, 2023
|
|
8ATD
 
 | | Wild type hexamer oxalyl-CoA synthetase (OCS) | | Descriptor: | Oxalate--CoA ligase | | Authors: | Lill, P, Burgi, J, Raunser, S, Wilmanns, M, Gatsogiannis, C. | | Deposit date: | 2022-08-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Asymmetric horseshoe-like assembly of peroxisomal yeast oxalyl-CoA synthetase.
Biol.Chem., 404, 2023
|
|
