6M5A
 
 | | Crystal structure of GH121 beta-L-arabinobiosidase HypBA2 from Bifidobacterium longum | | Descriptor: | 1,2-ETHANEDIOL, Beta-L-arabinobiosidase, CALCIUM ION, ... | | Authors: | Saito, K, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2020-03-10 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of beta-L-arabinobiosidase belonging to glycoside hydrolase family 121.
Plos One, 15, 2020
|
|
6L93
 
 | |
8K7Y
 
 | | Crystal structure of GH146 beta-L-arabinofuranosidase Bll3HypBA1 (amino acids 380-1051), ligand-free form | | Descriptor: | ZINC ION, beta1,3-L-arabinofuranoside | | Authors: | Maruyama, S, Pan, L, Miyake, M, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-07-27 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bifidobacterial GH146 beta-L-arabinofuranosidase for the removal of beta 1,3-L-arabinofuranosides on plant glycans.
Appl.Microbiol.Biotechnol., 108, 2024
|
|
8K7X
 
 | | Crystal structure of GH146 beta-L-arabinofuranosidase Bll3HypBA1 (amino acids 380-1223) in complex with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Pan, L, Maruyama, S, Miyake, M, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-07-27 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Bifidobacterial GH146 beta-L-arabinofuranosidase for the removal of beta 1,3-L-arabinofuranosides on plant glycans.
Appl.Microbiol.Biotechnol., 108, 2024
|
|
8IC1
 
 | | endo-alpha-D-arabinanase EndoMA1 D51N mutant from Microbacterium arabinogalactanolyticum in complex with arabinooligosaccharides | | Descriptor: | (3~{a}~{S},5~{R},6~{R},6~{a}~{S})-5-(hydroxymethyl)-2,2-dimethyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]dioxol-6-ol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Li, J, Nakashima, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
8IC7
 
 | | exo-beta-D-arabinofuranosidase ExoMA2 from Microbacterium arabinogalactanolyticum in complex with beta-D-arabinofuranose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Fukushima, R, Kashima, T, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
8IC6
 
 | | exo-beta-D-arabinanase ExoMA2 from Microbacterium arabinogalactanolyticum in complex with Tris | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Fukushima, R, Kashima, T, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
8IC8
 
 | | Exo-alpha-D-arabinofuranosidase from Microbacterium arabinogalactanolyticum | | Descriptor: | Exo-alpha-D-arabinofuranosidase, PHOSPHATE ION | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
7D5V
 
 | | Structure of the C646A mutant of peptidylarginine deiminase type III (PAD3) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein-arginine deiminase type-3 | | Authors: | Akimoto, M, Mashimo, R, Unno, M. | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structures of human peptidylarginine deiminase type III provide insights into substrate recognition and inhibitor design.
Arch.Biochem.Biophys., 708, 2021
|
|
7D56
 
 | |
7DAN
 
 | |
7D4Y
 
 | |
7D5R
 
 | | Structure of the Ca2+-bound C646A mutant of peptidylarginine deiminase type III (PAD3) | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Mashimo, R, Akimoto, M, Unno, M. | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.148 Å) | | Cite: | Structures of human peptidylarginine deiminase type III provide insights into substrate recognition and inhibitor design.
Arch.Biochem.Biophys., 708, 2021
|
|
7D8N
 
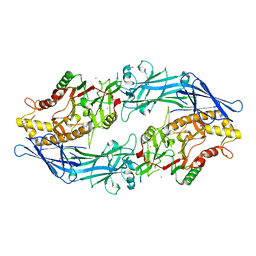 | | Structure of the inactive form of wild-type peptidylarginine deiminase type III (PAD3) crystallized under the condition with high concentrations of Ca2+ | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Funabashi, K, Sawata, M, Unno, M. | | Deposit date: | 2020-10-08 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Structures of human peptidylarginine deiminase type III provide insights into substrate recognition and inhibitor design.
Arch.Biochem.Biophys., 708, 2021
|
|
1QDQ
 
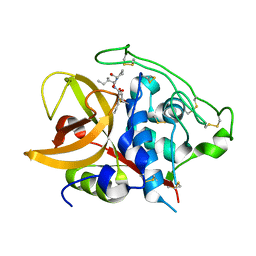 | | X-RAY CRYSTAL STRUCTURE OF BOVINE CATHEPSIN B-CA074 COMPLEX | | Descriptor: | CATHEPSIN B, [PROPYLAMINO-3-HYDROXY-BUTAN-1,4-DIONYL]-ISOLEUCYL-PROLINE | | Authors: | Yamamoto, A. | | Deposit date: | 1999-07-10 | | Release date: | 2000-07-10 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Substrate specificity of bovine cathepsin B and its inhibition by CA074, based on crystal structure refinement of the complex.
J.Biochem.(Tokyo), 127, 2000
|
|
7DEV
 
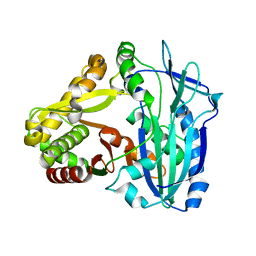 | | Crystal Structures of Anthocyanin 5,3'-aromatic acyltransferase from Gentiana triflora | | Descriptor: | Anthocyanin 5-aromatic acyltransferase | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2020-11-05 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Anthocyanin 5,3'-aromatic acyltransferase from Gentiana triflora, a structural insight into biosynthesis of a blue anthocyanin.
Phytochemistry, 186, 2021
|
|
7DEX
 
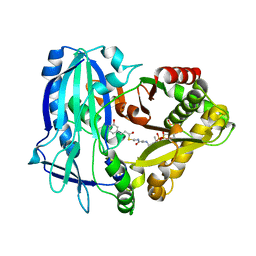 | | Crystal Structures of Anthocyanin 5,3'-aromatic acyltransferase H174A mutant with caffeoyl-CoA | | Descriptor: | Anthocyanin 5-aromatic acyltransferase, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] (E)-3-[3,4-bis(oxidanyl)phenyl]prop-2-enethioate | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2020-11-05 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Anthocyanin 5,3'-aromatic acyltransferase from Gentiana triflora, a structural insight into biosynthesis of a blue anthocyanin.
Phytochemistry, 186, 2021
|
|
5ZQQ
 
 | | Tankyrase-2 in complex with compound 52 | | Descriptor: | 1-methyl-1'-(4-oxo-3,4,5,6,7,8-hexahydroquinazolin-2-yl)spiro[indole-3,4'-piperidin]-2(1H)-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Yoshimoto, N, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2018-04-19 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of Novel Spiroindoline Derivatives as Selective Tankyrase Inhibitors.
J. Med. Chem., 62, 2019
|
|
5ZQO
 
 | | Tankyrase-2 in complex with compound 1a | | Descriptor: | 2-[4-(2-methoxyphenyl)piperazin-1-yl]-5,6,7,8-tetrahydroquinazolin-4(3H)-one, GLYCEROL, SULFATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Yoshimoto, N, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2018-04-19 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Discovery of Novel Spiroindoline Derivatives as Selective Tankyrase Inhibitors.
J. Med. Chem., 62, 2019
|
|
6KRO
 
 | | Tankyrase-2 in complex with RK-582 | | Descriptor: | 6-[(2S,6R)-2,6-dimethylmorpholin-4-yl]-4-fluoranyl-1-methyl-1'-(8-methyl-4-oxidanylidene-3,5,6,7-tetrahydropyrido[2,3-d]pyrimidin-2-yl)spiro[indole-3,4'-piperidine]-2-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2019-08-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and Discovery of an Orally Efficacious Spiroindolinone-Based Tankyrase Inhibitor for the Treatment of Colon Cancer.
J.Med.Chem., 63, 2020
|
|
5ZQP
 
 | | Tankyrase-2 in complex with compound 12 | | Descriptor: | 1'-(4-oxo-3,4,5,6,7,8-hexahydroquinazolin-2-yl)-2H-spiro[1-benzofuran-3,4'-piperidin]-2-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Yoshimoto, N, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2018-04-19 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of Novel Spiroindoline Derivatives as Selective Tankyrase Inhibitors.
J. Med. Chem., 62, 2019
|
|
5ZQR
 
 | | Tankyrase-2 in complex with compound 40c | | Descriptor: | 2-[4,6-difluoro-1-(2-hydroxyethyl)-1,2-dihydro-1'H-spiro[indole-3,4'-piperidin]-1'-yl]-5,6,7,8-tetrahydroquinazolin-4(3H)-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Yoshimoto, N, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2018-04-19 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Novel Spiroindoline Derivatives as Selective Tankyrase Inhibitors.
J. Med. Chem., 62, 2019
|
|
6A84
 
 | | Tankyrase-2 in complex with compound 15d | | Descriptor: | 2-(4-chloro-1,2-dihydro-1'H-spiro[indole-3,4'-piperidin]-1'-yl)-5,6,7,8-tetrahydroquinazolin-4(3H)-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Yoshimoto, N, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2018-07-06 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of Novel Spiroindoline Derivatives as Selective Tankyrase Inhibitors.
J. Med. Chem., 62, 2019
|
|
3NK4
 
 | |
1TEZ
 
 | | COMPLEX BETWEEN DNA AND THE DNA PHOTOLYASE FROM ANACYSTIS NIDULANS | | Descriptor: | 5'-D(*AP*TP*CP*GP*GP*CP*T*(TCP)P*CP*GP*C)-3', 5'-D(*TP*CP*GP*C)-3', 5'-D(P*CP*GP*AP*AP*GP*CP*CP*GP*A)-3', ... | | Authors: | Essen, L.-O, Carell, T, Mees, A, Klar, T. | | Deposit date: | 2004-05-26 | | Release date: | 2004-12-14 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair
Science, 306, 2004
|
|
