8JB5
 
 | |
7NEI
 
 | |
4RX5
 
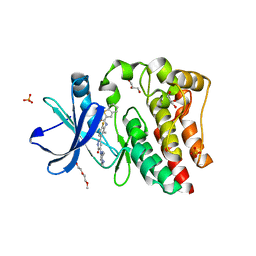 | | Bruton's tyrosine kinase (BTK) with pyridazinone compound 23 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, GLYCEROL, N-(6-fluoro-2-methyl-3-{5-[(5-methyl-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-2-yl)amino]-6-oxo-1,6-dihydropyridazin-3-yl}phenyl)-1-benzothiophene-2-carboxamide, ... | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2014-12-08 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.356 Å) | | Cite: | Discovery of highly potent and selective Bruton's tyrosine kinase inhibitors: Pyridazinone analogs with improved metabolic stability.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5HYZ
 
 | | Crystal Structure of SCL7 in Oryza sativa | | Descriptor: | GRAS family transcription factor containing protein, expressed | | Authors: | Wu, Y, Li, S, Zhao, Y, Sun, L. | | Deposit date: | 2016-02-02 | | Release date: | 2016-04-27 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Crystal Structure of the GRAS Domain of SCARECROW-LIKE7 in Oryza sativa.
Plant Cell, 28, 2016
|
|
5XGV
 
 | |
2P3U
 
 | |
2P3T
 
 | |
6ITP
 
 | | Crystal structure of cortisol complexed with its nanobody at pH 3.5 | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, anti-cortisol camelid antibody | | Authors: | Ding, Y, Ding, L.L, Wang, Z.Y, Zhong, P.Y. | | Deposit date: | 2018-11-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | Structural insights into the mechanism of single domain VHH antibody binding to cortisol.
Febs Lett., 593, 2019
|
|
6ITQ
 
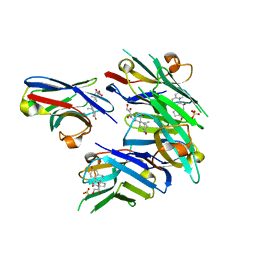 | | Crystal structure of cortisol complexed with its nanobody at pH 10.5 | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, SULFATE ION, anti-cortisol camelid antibody | | Authors: | Ding, Y, Ding, L.L, Wang, Z.Y, Zhong, P.Y. | | Deposit date: | 2018-11-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.526 Å) | | Cite: | Structural insights into the mechanism of single domain VHH antibody binding to cortisol.
Febs Lett., 593, 2019
|
|
2I7B
 
 | |
1QA0
 
 | | BOVINE TRYPSIN 2-AMINOBENZIMIDAZOLE COMPLEX | | Descriptor: | 2H-BENZOIMIDAZOL-2-YLAMINE, CALCIUM ION, TRYPSIN | | Authors: | Whitlow, M. | | Deposit date: | 1999-04-09 | | Release date: | 2000-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic analysis of potent and selective factor Xa inhibitors complexed to bovine trypsin.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4XUC
 
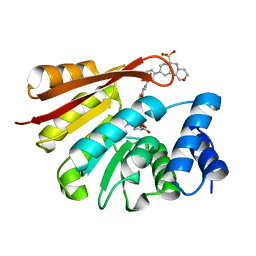 | | Synthesis and evaluation of heterocyclic catechol mimics as inhibitors of catechol-O-methyltransferase (COMT): Structure with Cmpd18 (1-(biphenyl-3-yl)-3-hydroxypyridin-4(1H)-one) | | Descriptor: | 1-(biphenyl-3-yl)-3-hydroxypyridin-4(1H)-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Catechol O-methyltransferase, ... | | Authors: | Allison, T, Wolkenberg, S, Sanders, J.M, Soisson, S.M. | | Deposit date: | 2015-01-25 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and Evaluation of Heterocyclic Catechol Mimics as Inhibitors of Catechol-O-methyltransferase (COMT).
Acs Med.Chem.Lett., 6, 2015
|
|
4XUD
 
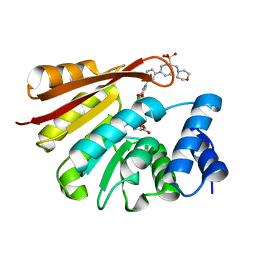 | | Synthesis and evaluation of heterocyclic catechol mimics as inhibitors of catechol-O-methyltransferase (COMT): Structure with Cmpd32 ([1-(biphenyl-3-yl)-5-hydroxy-4-oxo-1,4-dihydropyridin-3-yl]boronic acid) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Allison, T, Wolkenberg, S, Sanders, J.M, Soisson, S.M. | | Deposit date: | 2015-01-25 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and Evaluation of Heterocyclic Catechol Mimics as Inhibitors of Catechol-O-methyltransferase (COMT).
Acs Med.Chem.Lett., 6, 2015
|
|
8GRR
 
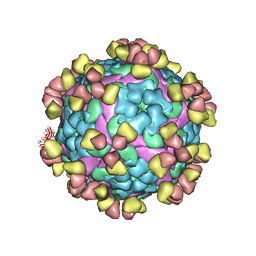 | | Complex of FMDV A/WH/CHA/09 and bovine neutralizing scFv antibody W125 | | Descriptor: | A/WH/CHA/09 VP1, A/WH/CHA/09 VP2, A/WH/CHA/09 VP3, ... | | Authors: | He, Y, Kun, L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Conserved antigen structures and antibody-driven variations on foot-and-mouth disease virus serotype A revealed by bovine neutralizing monoclonal antibodies.
Plos Pathog., 19, 2023
|
|
8GSP
 
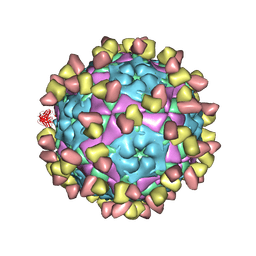 | | Complex of FMDV A/WH/CHA/09 and bovine neutralizing scFv antibody W2 | | Descriptor: | A/WH/CHA/09 VP1, A/WH/CHA/09 VP2, A/WH/CHA/09 VP3, ... | | Authors: | He, Y, Li, K. | | Deposit date: | 2022-09-06 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Conserved antigen structures and antibody-driven variations on foot-and-mouth disease virus serotype A revealed by bovine neutralizing monoclonal antibodies.
Plos Pathog., 19, 2023
|
|
4XUE
 
 | | Synthesis and evaluation of heterocyclic catechol mimics as inhibitors of catechol-O-methyltransferase (COMT): Structure with Cmpd27b | | Descriptor: | 2-(biphenyl-3-yl)-5-hydroxy-3-methylpyrimidin-4(3H)-one, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Allison, T, Wolkenberg, S, Sanders, J.M, Soisson, S.M. | | Deposit date: | 2015-01-25 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and Evaluation of Heterocyclic Catechol Mimics as Inhibitors of Catechol-O-methyltransferase (COMT).
Acs Med.Chem.Lett., 6, 2015
|
|
4Y7R
 
 | | Crystal structure of WDR5 in complex with MYC MbIIIb peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MYC MbIIIb peptide, ... | | Authors: | Sun, Q, Phan, J, Olejniczak, E.T, Thomas, L.R, Fesik, S.W, Tansey, W.P. | | Deposit date: | 2015-02-16 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Interaction with WDR5 Promotes Target Gene Recognition and Tumorigenesis by MYC.
Mol.Cell, 58, 2015
|
|
8H9I
 
 | | Human ATP synthase F1 domain, state2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit O, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
8H9Q
 
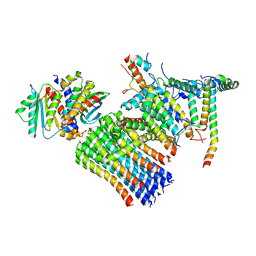 | | Human ATP synthase state 3b subregion 3 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase F(0) complex subunit C1, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
8H9R
 
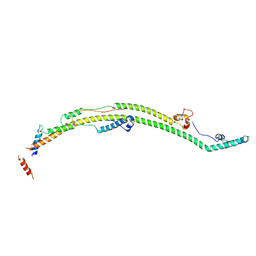 | | Human ATP synthase state 3b subregion 2 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase subunit d, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
8H9P
 
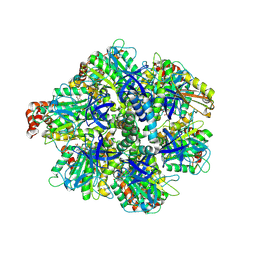 | | Human ATP synthase F1 domain, state 3b | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit O, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
8H9U
 
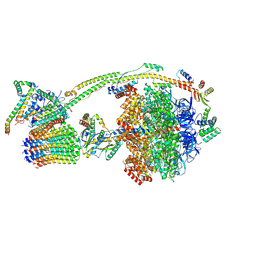 | | Human ATP synthase state 3a (combined) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F(0) complex subunit B1, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
8H9V
 
 | | Human ATP synthase state 3b (combined) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F(0) complex subunit B1, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
8H9M
 
 | | Human ATP synthase state 3a subregion 3 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase F(0) complex subunit C1, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
8WDS
 
 | | Crystal structure of BF.7 RBD complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, W, Xie, Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Key mechanistic features of the trade-off between antibody escape and host cell binding in the SARS-CoV-2 Omicron variant spike proteins.
Embo J., 43, 2024
|
|
