4RO5
 
 | | Crystal structure of the SAT domain from the non-reducing fungal polyketide synthase CazM | | Descriptor: | GLYCEROL, SAT domain from CazM | | Authors: | Winter, J.M, Cascio, D, Sawaya, M.R, Tang, Y. | | Deposit date: | 2014-10-27 | | Release date: | 2015-09-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and Structural Basis for Controlling Chemical Modularity in Fungal Polyketide Biosynthesis.
J.Am.Chem.Soc., 137, 2015
|
|
4RPM
 
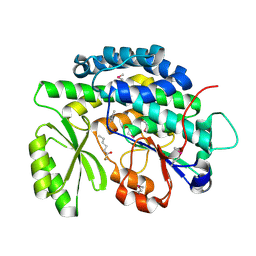 | | Crystal structure of the SAT domain from the non-reducing fungal polyketide synthase CazM with bound hexanoyl | | Descriptor: | HEXANOIC ACID, HEXANOYL-COENZYME A, SAT domain from CazM | | Authors: | Winter, J.M, Cascio, D, Sawaya, M.R, Tang, Y. | | Deposit date: | 2014-10-30 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and Structural Basis for Controlling Chemical Modularity in Fungal Polyketide Biosynthesis.
J.Am.Chem.Soc., 137, 2015
|
|
3W36
 
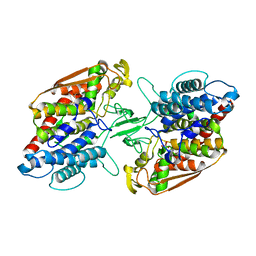 | | Crystal structure of holo-type bacterial Vanadium-dependent chloroperoxidase | | Descriptor: | NapH1, VANADATE ION | | Authors: | Liscombe, D.K, Miyanaga, A, Fielding, E, Bernhardt, P, Li, A, Winter, J.M, Gilson, M.K, Noel, J.P, Moore, B.S. | | Deposit date: | 2012-12-11 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis of Stereospecific Vanadium-Dependent Haloperoxidase Family Enzymes in Napyradiomycin Biosynthesis.
Biochemistry, 2022
|
|
3W35
 
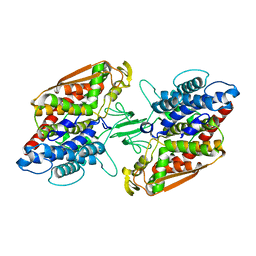 | | Crystal structure of apo-type bacterial Vanadium-dependent chloroperoxidase | | Descriptor: | NapH1 | | Authors: | Liscombe, D.K, Miyanaga, A, Fielding, E, Bernhardt, P, Li, A, Winter, J.M, Gilson, M.K, Noel, J.P, Moore, B.S. | | Deposit date: | 2012-12-11 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Stereospecific Vanadium-Dependent Haloperoxidase Family Enzymes in Napyradiomycin Biosynthesis.
Biochemistry, 2022
|
|
8PWH
 
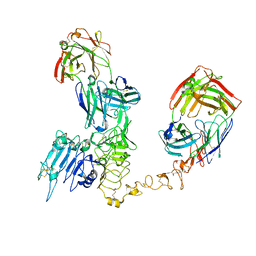 | | Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pertuzumab Fab heavy chain, Pertuzumab Fab light chain, ... | | Authors: | Ruedas, R, Vuillemot, R, Tubiana, T, Winter, J.M, Pieri, L, Arteni, A.A, Samson, C, Jonic, J, Mathieu, M, Bressanelli, S. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structure and conformational variability of the HER2-trastuzumab-pertuzumab complex.
J.Struct.Biol., 216, 2024
|
|
8Q6J
 
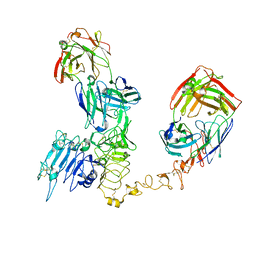 | | Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pertuzumab Fab heavy chain, Pertuzumab Fab light chain, ... | | Authors: | Ruedas, R, Vuillemot, R, Tubiana, T, Winter, J.M, Pieri, L, Arteni, A.A, Samson, C, Jonic, J, Mathieu, M, Bressanelli, S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and conformational variability of the HER2-trastuzumab-pertuzumab complex.
J.Struct.Biol., 216, 2024
|
|
7LUB
 
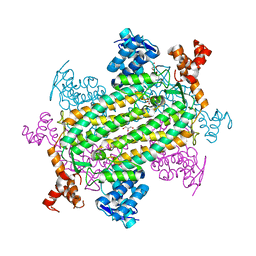 | |
7MBH
 
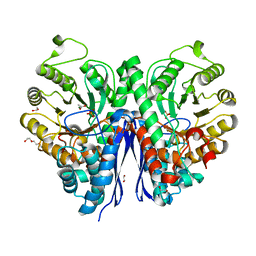 | | Structure of Human Enolase 2 in complex with phosphoserine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Gamma-enolase, ... | | Authors: | Leonard, P.G, Hicks, K.G, Rutter, J. | | Deposit date: | 2021-03-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein-metabolite interactomics of carbohydrate metabolism reveal regulation of lactate dehydrogenase.
Science, 379, 2023
|
|
8CXL
 
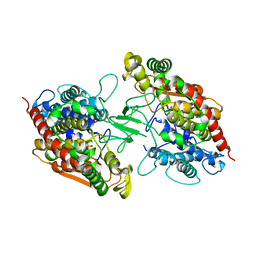 | | Structure of NapH3, a vanadium-dependent haloperoxidase homolog catalyzing the stereospecific alpha-hydroxyketone rearrangement reaction in napyradiomycin biosynthesis | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NapH3 | | Authors: | Chen, P.Y.-T, Chekan, J.R, Moore, B.S. | | Deposit date: | 2022-05-21 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Basis of Stereospecific Vanadium-Dependent Haloperoxidase Family Enzymes in Napyradiomycin Biosynthesis.
Biochemistry, 61, 2022
|
|
8A50
 
 | | Crystal structure of HSF2BP-ALPHA1 tetramer | | Descriptor: | Heat shock factor 2-binding protein, PHOSPHATE ION | | Authors: | Miron, S, Legrand, P, Ropars, V, Ghouil, R, Zinn-Justin, S. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | BRCA2-HSF2BP oligomeric ring disassembly by BRME1 promotes homologous recombination.
Sci Adv, 9, 2023
|
|
8A51
 
 | | Crystal structure of HSF2BP-BRME1 complex | | Descriptor: | 1,2-ETHANEDIOL, Break repair meiotic recombinase recruitment factor 1, CHLORIDE ION, ... | | Authors: | Miron, S, Legrand, P, Ropars, V, Ghouil, R, Zinn-Justin, S. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | BRCA2-HSF2BP oligomeric ring disassembly by BRME1 promotes homologous recombination.
Sci Adv, 9, 2023
|
|
7Q5G
 
 | | LAN-DAP5 DERIVATIVE OF LANREOTIDE: L-DIAMINO PROPIONIC ACID IN POSITION 5 IN PLACE OF L-LYSINE | | Descriptor: | ETHANOL, LAN-DAP5 DERIVATIVE OF LANREOTIDE | | Authors: | Bressanelli, S, Le Du, M.H, Gobeaux, F, Legrand, P, Paternostre, M. | | Deposit date: | 2021-11-03 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | Atomic structure of Lanreotide nanotubes revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Q5A
 
 | | Lanreotide nanotube | | Descriptor: | Lanreotide | | Authors: | Pieri, L, Wang, F, Arteni, A.A, Bressanelli, S, Egelman, E.H, Paternostre, M. | | Deposit date: | 2021-11-03 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Atomic structure of Lanreotide nanotubes revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RO4
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 3 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7RO2
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 1 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7RO5
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 4 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7ROB
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 6 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7RO6
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 5 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7RO3
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 2 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7ROI
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 12 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7ROH
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 11 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7ROD
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 8 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7ROE
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 9 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7ROG
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 10 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7ROC
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 7 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
