8E6K
 
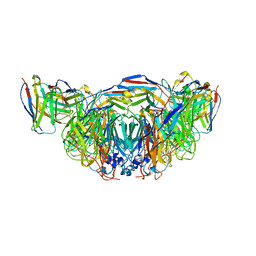 | | 2H08 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2H08 fragment antigen binding heavy chain, 2H08 fragment antigen binding light chain, ... | | Authors: | Turner, H.L, Ozorowski, G, Ward, A.B. | | Deposit date: | 2022-08-22 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Human anti-N1 monoclonal antibodies elicited by pandemic H1N1 virus infection broadly inhibit HxN1 viruses in vitro and in vivo.
Immunity, 56, 2023
|
|
8E6J
 
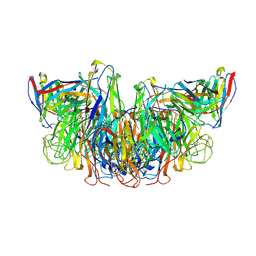 | |
6MLM
 
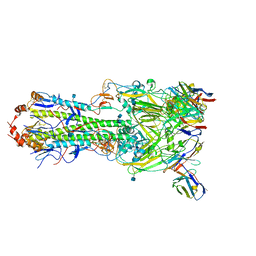 | | H7 HA0 in complex with Fv from H7.5 IgG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain Fv of H7.5 Fab, Hemagglutinin HA1 chain, ... | | Authors: | Pallesen, J, Turner, H.L, Ward, A.B. | | Deposit date: | 2018-09-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Potent anti-influenza H7 human monoclonal antibody induces separation of hemagglutinin receptor-binding head domains.
PLoS Biol., 17, 2019
|
|
6DID
 
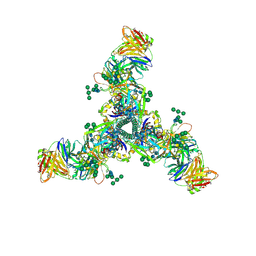 | | HIV Env BG505 SOSIP with polyclonal Fabs from immunized rabbit #3417 post-boost#1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Turner, H.L, Cottrell, C.A, Oyen, D, Wilson, I.A, Ward, A.B. | | Deposit date: | 2018-05-23 | | Release date: | 2018-09-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.71 Å) | | Cite: | Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization.
Immunity, 49, 2018
|
|
7JJI
 
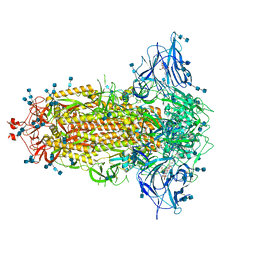 | | Structure of SARS-CoV-2 3Q-2P full-length prefusion spike trimer (C3 symmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-hydroxyethyl 2-deoxy-3,5-bis-O-(2-hydroxyethyl)-6-O-(2-{[(9E)-octadec-9-enoyl]oxy}ethyl)-alpha-L-xylo-hexofuranoside, ... | | Authors: | Bangaru, S, Turner, H.L, Ozorowski, G, Antanasijevic, A, Ward, A.B. | | Deposit date: | 2020-07-26 | | Release date: | 2020-08-26 | | Last modified: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate.
Science, 370, 2020
|
|
7JJJ
 
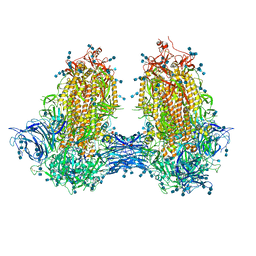 | | Structure of SARS-CoV-2 3Q-2P full-length dimers of spike trimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bangaru, S, Turner, H.L, Ozorowski, G, Antanasijevic, A, Ward, A.B. | | Deposit date: | 2020-07-26 | | Release date: | 2020-08-26 | | Last modified: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate.
Science, 370, 2020
|
|
6U02
 
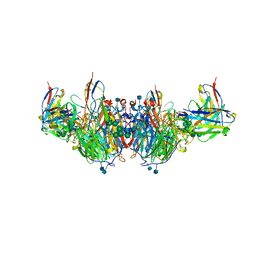 | | CryoEM-derived model of NA-63 Fab in complex with N9 Shanghai2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab-63 Heavy Chain, Fab-63 Light Chain, ... | | Authors: | Ward, A.B, Turner, H.L, Zhu, X. | | Deposit date: | 2019-08-13 | | Release date: | 2019-12-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
5KEL
 
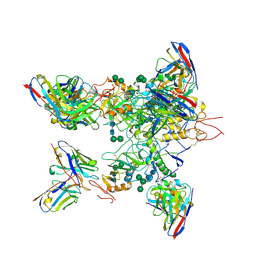 | | EBOV GP in complex with variable Fab domains of IgGs c2G4 and c13C6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ebola surface glycoprotein, ... | | Authors: | Pallesen, J, Murin, C.D, de Val, N, Cottrell, C.A, Hastie, K.M, Turner, H.L, Fusco, M.L, Flyak, A.I, Zeitlin, L, Crowe Jr, J.E, Andersen, K.G, Saphire, E.O, Ward, A.B. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structures of Ebola virus GP and sGP in complex with therapeutic antibodies.
Nat Microbiol, 1, 2016
|
|
5KEN
 
 | | EBOV GP in complex with variable Fab domains of IgGs c4G7 and c13C6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ebola surface glycoprotein, ... | | Authors: | Pallesen, J, Murin, C.D, de Val, N, Cottrell, C.A, Hastie, K.M, Turner, H.L, Fusco, M.L, Flyak, A.I, Zeitlin, L, Crowe Jr, J.E, Andersen, K.G, Saphire, E.O, Ward, A.B. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structures of Ebola virus GP and sGP in complex with therapeutic antibodies.
Nat Microbiol, 1, 2016
|
|
5KEM
 
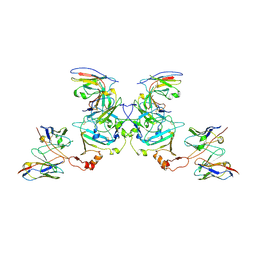 | | EBOV sGP in complex with variable Fab domains of IgGs c13C6 and BDBV91 | | Descriptor: | BDBV91 variable Fab domain heavy chain, BDBV91 variable Fab domain light chain, Ebola secreted glycoprotein, ... | | Authors: | Pallesen, J, Murin, C.D, de Val, N, Cottrell, C.A, Hastie, K.M, Turner, H.L, Fusco, M.L, Flyak, A.I, Zeitlin, L, Crowe Jr, J.E, Andersen, K.G, Saphire, E.O, Ward, A.B. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structures of Ebola virus GP and sGP in complex with therapeutic antibodies.
Nat Microbiol, 1, 2016
|
|
5I08
 
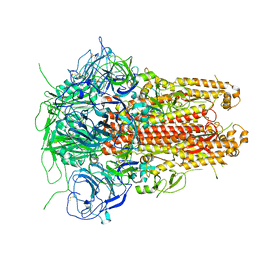 | | Prefusion structure of a human coronavirus spike protein | | Descriptor: | Spike glycoprotein,Foldon chimera | | Authors: | Kirchdoerfer, R.N, Cottrell, C.A, Wang, N, Pallesen, J, Yassine, H.M, Turner, H.L, Corbett, K.S, Graham, B.S, McLellan, J.S, Ward, A.B. | | Deposit date: | 2016-02-03 | | Release date: | 2016-03-02 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Pre-fusion structure of a human coronavirus spike protein.
Nature, 531, 2016
|
|
8DT8
 
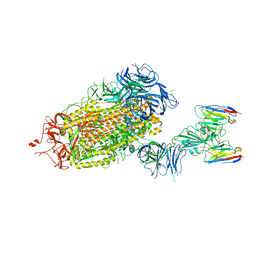 | | LM18/Nb136 bispecific tetra-nanobody immunoglobulin in complex with SARS-CoV-2-6P-Mut7 S protein (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LM18 nanobody, Nb136 nanobody, ... | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2022-07-25 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Fully synthetic platform to rapidly generate tetravalent bispecific nanobody-based immunoglobulins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7RU1
 
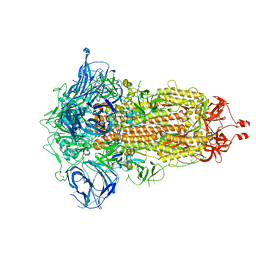 | | SARS-CoV-2-6P-Mut7 S protein (C3 symmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
7RU8
 
 | | CC6.30 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (RBD/Fv local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC6.30 Fab Kappa chain Fv, ... | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
7RU4
 
 | | CC6.33 IgG in complex with SARS-CoV-2-6P-Mut7 S protein (RBD/Fv local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CC6.33 IgG heavy chain Fv, ... | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
7RU3
 
 | | CC6.33 IgG in complex with SARS-CoV-2-6P-Mut7 S protein (non-uniform refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
7RU5
 
 | | CC6.30 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (non-uniform refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC6.30 Fab heavy chain Fv, ... | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
7RU2
 
 | | SARS-CoV-2-6P-Mut7 S protein (asymmetric) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
6PZW
 
 | | CryoEM derived model of NA-22 Fab in complex with N9 Shanghai2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NA-22 fragment antigen binding heavy chain, ... | | Authors: | Ward, A.B, Turner, H.L, Zhu, X. | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
6PZZ
 
 | | CryoEM derived model of NA-80 Fab in complex with N9 Shanghai2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NA-80 fragment antibody heavy chain, ... | | Authors: | Ward, A.B, Turner, H.L, Zhu, X. | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
6PZY
 
 | | CryoEM derived model of NA-73 Fab in complex with N9 Shanghai2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NA-73 fragment antibody heavy chain, ... | | Authors: | Ward, A.B, Turner, H.L, Zhu, X. | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
6CRZ
 
 | | SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CS2
 
 | | SARS Spike Glycoprotein - human ACE2 complex, Stabilized variant, all ACE2-bound particles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CS1
 
 | | SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, two S1 CTDs in an upwards conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CRX
 
 | | SARS Spike Glycoprotein, Stabilized variant, two S1 CTDs in the upwards conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
