6ULG
 
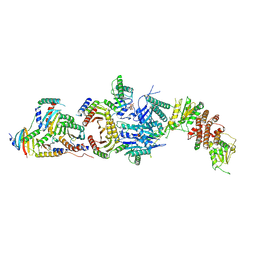 | | Cryo-EM structure of the FLCN-FNIP2-Rag-Ragulator complex | | Descriptor: | Folliculin, Folliculin-interacting protein 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Shen, K, Rogala, K.B, Yu, Z.H, Sabatini, D.M. | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Cryo-EM Structure of the Human FLCN-FNIP2-Rag-Ragulator Complex.
Cell, 179, 2019
|
|
6CES
 
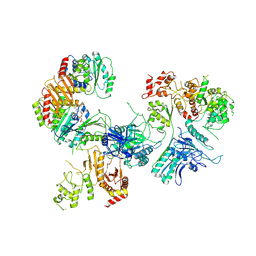 | | Cryo-EM structure of GATOR1-RAG | | Descriptor: | GATOR complex protein DEPDC5, GATOR complex protein NPRL2, GATOR complex protein NPRL3, ... | | Authors: | Shen, K, Huang, R.K, Brignole, E.J, Yu, Z, Sabatini, D.M. | | Deposit date: | 2018-02-12 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Architecture of the human GATOR1 and GATOR1-Rag GTPases complexes.
Nature, 556, 2018
|
|
6CET
 
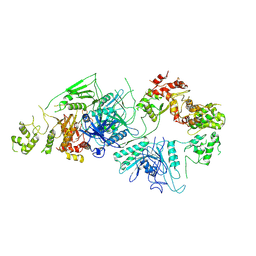 | | Cryo-EM structure of GATOR1 | | Descriptor: | GATOR complex protein DEPDC5, GATOR complex protein NPRL2, GATOR complex protein NPRL3 | | Authors: | Shen, K, Huang, R.K, Brignole, E.J, Yu, Z, Sabatini, D.M. | | Deposit date: | 2018-02-12 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Architecture of the human GATOR1 and GATOR1-Rag GTPases complexes.
Nature, 556, 2018
|
|
9DYG
 
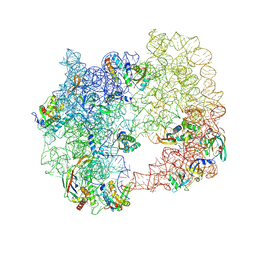 | | iSAT-PNA-RA20, RA20-B-c class | | Descriptor: | 23S rRNA, Large ribosomal subunit protein bL17, Large ribosomal subunit protein bL19, ... | | Authors: | Sheng, K, Dong, X, Lee, J. | | Deposit date: | 2024-10-14 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.27 Å) | | Cite: | Domain consolidation in bacterial 50S assembly revealed by anti-sense oligonucleotide probing
To Be Published
|
|
6XS6
 
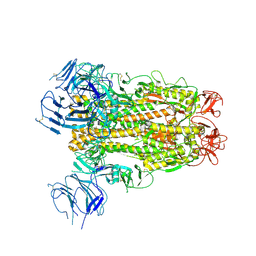 | | SARS-CoV-2 Spike D614G variant, minus RBD | | Descriptor: | Spike glycoprotein | | Authors: | Wang, X, Egri, S.B, Dudkina, N, Luban, J, Shen, K. | | Deposit date: | 2020-07-15 | | Release date: | 2020-07-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant.
Cell, 183, 2020
|
|
7T3A
 
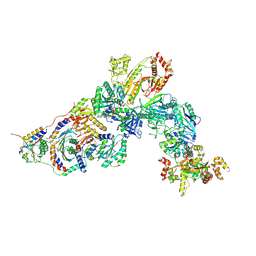 | | GATOR1-RAG-RAGULATOR - Inhibitory Complex | | Descriptor: | GATOR complex protein DEPDC5, GATOR complex protein NPRL2, GATOR complex protein NPRL3, ... | | Authors: | Egri, S.B, Shen, K. | | Deposit date: | 2021-12-07 | | Release date: | 2022-04-06 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of the human GATOR1-Rag-Ragulator complex reveal a spatial-constraint regulated GAP mechanism.
Mol.Cell, 82, 2022
|
|
7T3B
 
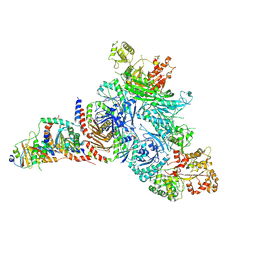 | | GATOR1-RAG-RAGULATOR - GAP Complex | | Descriptor: | ALUMINUM FLUORIDE, GATOR complex protein DEPDC5, GATOR complex protein NPRL2, ... | | Authors: | Egri, S.B, Shen, K. | | Deposit date: | 2021-12-07 | | Release date: | 2022-04-06 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of the human GATOR1-Rag-Ragulator complex reveal a spatial-constraint regulated GAP mechanism.
Mol.Cell, 82, 2022
|
|
7T3C
 
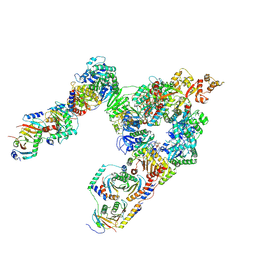 | | GATOR1-RAG-RAGULATOR - Dual Complex | | Descriptor: | ALUMINUM FLUORIDE, GATOR complex protein DEPDC5, GATOR complex protein NPRL2, ... | | Authors: | Egri, S.B, Shen, K. | | Deposit date: | 2021-12-07 | | Release date: | 2022-04-06 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of the human GATOR1-Rag-Ragulator complex reveal a spatial-constraint regulated GAP mechanism.
Mol.Cell, 82, 2022
|
|
5VOK
 
 | | Crystal structure of the C7orf59-HBXIP dimer | | Descriptor: | Ragulator complex protein LAMTOR4, Ragulator complex protein LAMTOR5 | | Authors: | Rasheed, N, Nascimento, A.F.Z, Bar-Peled, L, Shen, K, Sabatini, D.M, Aparicio, R, Smetana, J.H.C. | | Deposit date: | 2017-05-03 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | C7orf59/Lamtor4 phosphorylation and structural flexibility modulate Ragulator assembly.
Febs Open Bio, 2019
|
|
4C7O
 
 | | The structural basis of FtsY recruitment and GTPase activation by SRP RNA | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SIGNAL RECOGNITION PARTICLE PROTEIN, ... | | Authors: | Voigts-Hoffmann, F, Schmitz, N, Shen, K, Shan, S.O, Ataide, S.F, Ban, N. | | Deposit date: | 2013-09-23 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structural Basis of Ftsy Recruitment and Gtpase Activation by Srp RNA
Mol.Cell, 52, 2013
|
|
2CCI
 
 | | Crystal structure of phospho-CDK2 Cyclin A in complex with a peptide containing both the substrate and recruitment sites of CDC6 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6 homolog, Cyclin-A2, ... | | Authors: | Cheng, K.Y, Noble, M.E.M, Skamnaki, V, Brown, N.R, Lowe, E.D, Kontogiannis, L, Shen, K, Cole, P.A, Siligardi, G, Johnson, L.N. | | Deposit date: | 2006-01-16 | | Release date: | 2006-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of the phospho-CDK2/cyclin A recruitment site in substrate recognition.
J. Biol. Chem., 281, 2006
|
|
2CCH
 
 | | The crystal structure of CDK2 cyclin A in complex with a substrate peptide derived from CDC modified with a gamma-linked ATP analogue | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION CONTROL PROTEIN 6 HOMOLOG, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Cheng, K.Y, Noble, M.E.M, Skamnaki, V, Brown, N.R, Lowe, E.D, Kontogiannis, L, Shen, K, Cole, P.A, Siligardi, G, Johnson, L.N. | | Deposit date: | 2006-01-16 | | Release date: | 2006-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Role of the Phospho-Cdk2/Cyclin a Recruitment Site in Substrate Recognition
J.Biol.Chem., 281, 2006
|
|
1PXH
 
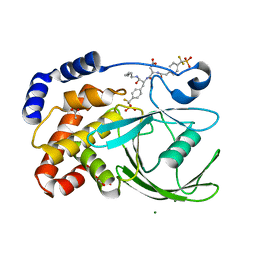 | | Crystal structure of protein tyrosine phosphatase 1B with potent and selective bidentate inhibitor compound 2 | | Descriptor: | ACETIC ACID, MAGNESIUM ION, N-{1-[5-(1-CARBAMOYL-2-MERCAPTO-ETHYLCARBAMOYL)-PENTYLCARBAMOYL]-2-[4-(DIFLUORO-PHOSPHONO-METHYL)-PHENYL]-ETHYL}-3-{2-[4-(DIFLUORO-PHOSPHONO-METHYL)-PHENYL]-ACETYLAMINO}-SUCCINAMIC ACID, ... | | Authors: | Sun, J.P, Fedorov, A, Lee, S.Y, Guo, X.L, Shen, K, Lawrence, D.S, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2003-07-04 | | Release date: | 2003-08-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of PTP1B complexed with a potent and selective bidentate inhibitor.
J.Biol.Chem., 278, 2003
|
|
2XXA
 
 | | The Crystal Structure of the Signal Recognition Particle (SRP) in Complex with its Receptor(SR) | | Descriptor: | 4.5S RNA, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Ataide, S.F, Schmitz, N, Shen, K, Ke, A, Shan, S, Doudna, J.A, Ban, N. | | Deposit date: | 2010-11-09 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | The Crystal Structure of the Signal Recognition Particle in Complex with its Receptor.
Science, 331, 2011
|
|
8FW5
 
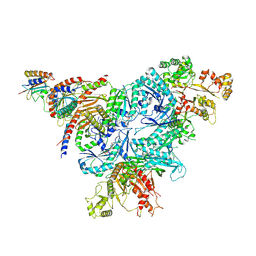 | | Chimeric HsGATOR1-SpGtr-SpLam complex | | Descriptor: | ALUMINUM FLUORIDE, GATOR complex protein DEPDC5, GATOR complex protein NPRL2, ... | | Authors: | Tettoni, S.D, Egri, S.B, Doxsey, D.D, Ouch, C, Chang, J, Song, K, Xu, C, Shen, K. | | Deposit date: | 2023-01-20 | | Release date: | 2023-07-26 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure of the Schizosaccharomyces pombe Gtr-Lam complex reveals evolutionary divergence of mTORC1-dependent amino acid sensing.
Structure, 31, 2023
|
|
8GB0
 
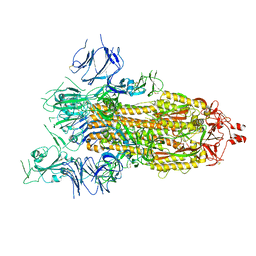 | |
2GS7
 
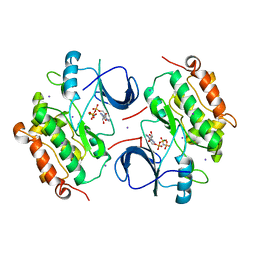 | | Crystal Structure of the inactive EGFR kinase domain in complex with AMP-PNP | | Descriptor: | Epidermal growth factor receptor, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Zhang, X, Gureasko, J, Shen, K, Cole, P.A, Kuriyan, J. | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor
Cell(Cambridge,Mass.), 125, 2006
|
|
2GS2
 
 | | Crystal Structure of the active EGFR kinase domain | | Descriptor: | Epidermal growth factor receptor | | Authors: | Zhang, X, Gureasko, J, Shen, K, Cole, P.A, Kuriyan, J. | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor.
Cell(Cambridge,Mass.), 125, 2006
|
|
2GS6
 
 | | Crystal Structure of the active EGFR kinase domain in complex with an ATP analog-peptide conjugate | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, Peptide, ... | | Authors: | Zhang, X, Gureasko, J, Shen, K, Cole, P.A, Kuriyan, J. | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An Allosteric Mechanism for Activation of the Kinase Domain of Epidermal Growth Factor Receptor
Cell(Cambridge,Mass.), 125, 2006
|
|
9CL9
 
 | | WT 12C IM fraction, B-b3 with RluB bound | | Descriptor: | 23S rRNA, Large ribosomal subunit protein bL20, Large ribosomal subunit protein bL21, ... | | Authors: | Lee, J, Sheng, K, Williamson, J.R. | | Deposit date: | 2024-07-10 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (5.04 Å) | | Cite: | 50S ribosome assembly intermediates at low temperature reveal bound RluB
To Be Published
|
|
6CSM
 
 | | Crystal structure of the natural light-gated anion channel GtACR1 | | Descriptor: | GtACR1, OLEIC ACID, RETINAL | | Authors: | Kato, H.E, Kim, Y, Yamashita, K, Kobilka, B.K, Deisseroth, K. | | Deposit date: | 2018-03-21 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural mechanisms of selectivity and gating in anion channelrhodopsins.
Nature, 561, 2018
|
|
6CSN
 
 | | Crystal structure of the designed light-gated anion channel iC++ at pH8.5 | | Descriptor: | CHLORIDE ION, OLEIC ACID, RETINAL, ... | | Authors: | Kato, H.E, Kim, Y, Yamashita, K, Kobilka, B.K, Deisseroth, K. | | Deposit date: | 2018-03-21 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural mechanisms of selectivity and gating in anion channelrhodopsins.
Nature, 561, 2018
|
|
6CSO
 
 | | Crystal structure of the designed light-gated anion channel iC++ at pH6.5 | | Descriptor: | OLEIC ACID, RETINAL, iC++ | | Authors: | Kato, H.E, Kim, Y, Yamashita, K, Kobilka, B.K, Deisseroth, K. | | Deposit date: | 2018-03-21 | | Release date: | 2018-09-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural mechanisms of selectivity and gating in anion channelrhodopsins.
Nature, 561, 2018
|
|
4FQ7
 
 | | Crystal structure of the maleate isomerase Iso from Pseudomonas putida S16 | | Descriptor: | Maleate cis-trans isomerase | | Authors: | Lu, Y, Chen, D, Zhang, Z, Li, Q, Wu, G, Xu, P. | | Deposit date: | 2012-06-25 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and computational studies of the maleate isomerase from Pseudomonas putida S16 reveal a breathing motion wrapping the substrate inside.
Mol.Microbiol., 87, 2013
|
|
4FQ5
 
 | | Crystal structure of the maleate isomerase Iso(C200A) from Pseudomonas putida S16 with maleate | | Descriptor: | MALEIC ACID, Maleate cis-trans isomerase | | Authors: | Lu, Y, Chen, D, Zhang, Z, Li, Q, Wu, G, Xu, P. | | Deposit date: | 2012-06-25 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and computational studies of the maleate isomerase from Pseudomonas putida S16 reveal a breathing motion wrapping the substrate inside.
Mol.Microbiol., 87, 2013
|
|
