7SS5
 
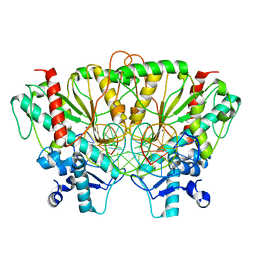 | |
5YI8
 
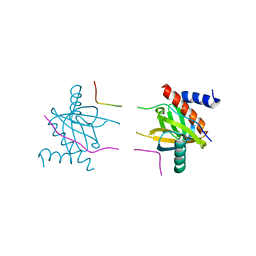 | |
5YI7
 
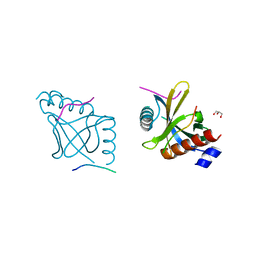 | |
8HND
 
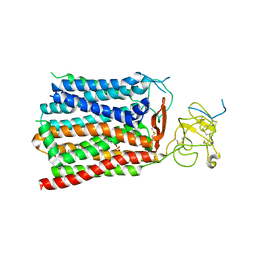 | | Cryo-EM structure of human OATP1B1 in complex with estrone-3-sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier organic anion transporter family member 1B1, ... | | Authors: | Shan, Z, Yang, X, Liu, H, Nan, J, Yuan, Y, Zhang, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
8HNH
 
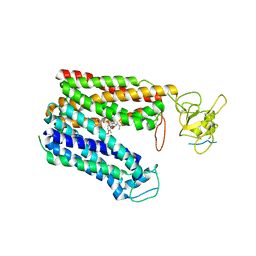 | | Cryo-EM structure of human OATP1B1 in complex with simeprevir | | Descriptor: | (2R,3aR,10Z,11aS,12aR,14aR)-N-(cyclopropylsulfonyl)-2-({7-methoxy-8-methyl-2-[4-(1-methylethyl)-1,3-thiazol-2-yl]quinolin-4-yl}oxy)-5-methyl-4,14-dioxo-2,3,3a,4,5,6,7,8,9,11a,12,13,14,14a-tetradecahydrocyclopenta[c]cyclopropa[g][1,6]diazacyclotetradecine-12a(1H)-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shan, Z, Yang, X, Zhang, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
8HNB
 
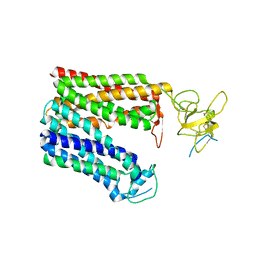 | | Cryo-EM structure of human OATP1B1 in apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier organic anion transporter family member 1B1 | | Authors: | Shan, Z, Yang, X, Zhang, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
8HNC
 
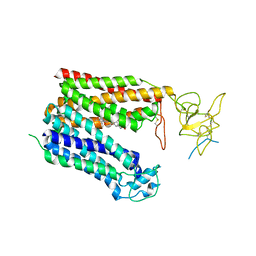 | | Cryo-EM structure of human OATP1B1 in complex with bilirubin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Shan, Z, Yang, X, Liu, H, Nan, J, Yuan, Y, Zhang, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
8K6L
 
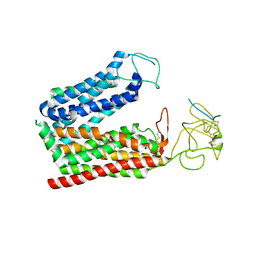 | | Cryo-EM structure of human OATP1B1 in complex with DCF | | Descriptor: | 2',7'-bis(chloranyl)-3',6'-bis(oxidanyl)spiro[2-benzofuran-3,9'-xanthene]-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shan, Z, Yang, X, Zhang, Y. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
3J8X
 
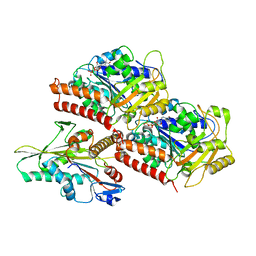 | | High-resolution structure of no-nucleotide kinesin on microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Shang, Z, Zhou, K, Xu, C, Csencsits, R, Cochran, J.C, Sindelar, C.V. | | Deposit date: | 2014-11-20 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation.
Elife, 3, 2014
|
|
3J8Y
 
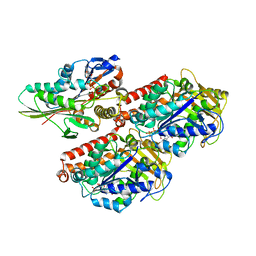 | | High-resolution structure of ATP analog-bound kinesin on microtubules | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shang, Z, Zhou, K, Xu, C, Csencsits, R, Cochran, J.C, Sindelar, C.V. | | Deposit date: | 2014-11-20 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation.
Elife, 3, 2014
|
|
5YGH
 
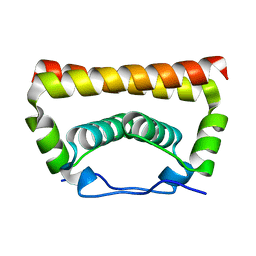 | | Crystal Structure of the Capsid Protein from Zika Virus | | Descriptor: | Capsid protein | | Authors: | Shang, Z, Song, H, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-09-23 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.884 Å) | | Cite: | Crystal Structure of the Capsid Protein from Zika Virus.
J. Mol. Biol., 430, 2018
|
|
5XMC
 
 | |
5CFF
 
 | | Crystal structure of Miranda/Staufen dsRBD5 complex | | Descriptor: | Miranda, Staufen | | Authors: | Shan, Z, Wen, W. | | Deposit date: | 2015-07-08 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural basis of Miranda-mediated Staufen localization during Drosophila neuroblast asymmetric division
Nat Commun, 6, 2015
|
|
7U32
 
 | | MVV cleaved synaptic complex (CSC) intasome at 3.4 A resolution | | Descriptor: | CALCIUM ION, DNA EV272, DNA EV273, ... | | Authors: | Shan, Z, Pye, V.E, Cherepanov, P, Lyumkis, D. | | Deposit date: | 2022-02-25 | | Release date: | 2022-05-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Multivalent interactions essential for lentiviral integrase function.
Nat Commun, 13, 2022
|
|
8SY7
 
 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound the unnatural dB-STP base pair in the active site | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shan, Z, Lyumkis, D, Oh, J, Wang, D. | | Deposit date: | 2023-05-24 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase.
Nat Commun, 14, 2023
|
|
8SY5
 
 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound the unnatural dS-BTP base pair in the active site | | Descriptor: | 2-oxo-2-hydroadenosine 5'-(tetrahydrogen triphosphate), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Shan, Z, Lyumkis, D, Oh, J, Wang, D. | | Deposit date: | 2023-05-24 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase.
Nat Commun, 14, 2023
|
|
8SY6
 
 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound the unnatural dB-UTP base pair in the active site | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Shan, Z, Lyumkis, D, Oh, J, Wang, D. | | Deposit date: | 2023-05-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase.
Nat Commun, 14, 2023
|
|
6J9R
 
 | |
5J19
 
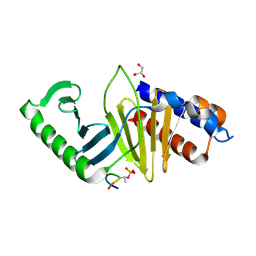 | | phospho-Pon binding-induced Plk1 dimerization | | Descriptor: | GLYCEROL, Phosphorylated peptide from Partner of Numb, Serine/threonine-protein kinase PLK1 | | Authors: | Zhu, K, Shan, Z, Zhang, L, Wen, W. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phospho-Pon Binding-Mediated Fine-Tuning of Plk1 Activity
Structure, 24, 2016
|
|
8TXO
 
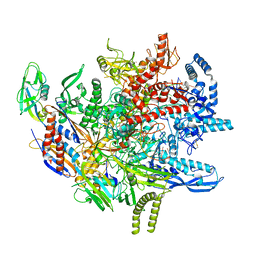 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound to the unnatural dZ-PTP base pair in the active site | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Aiyer, S, Shan, Z, Oh, J, Wang, D, Tan, Y.Z, Lyumkis, D. | | Deposit date: | 2023-08-23 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Overcoming resolution attenuation during tilted cryo-EM data collection.
Nat Commun, 15, 2024
|
|
7Y6I
 
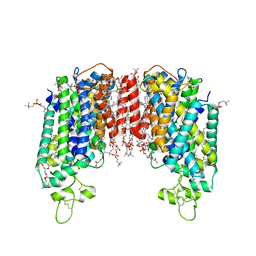 | | Cryo-EM structure of human sodium-chloride cotransporter | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nan, J, Yang, X, Shan, Z, Yuan, Y, Zhang, Y.Q. | | Deposit date: | 2022-06-20 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure of the human sodium-chloride cotransporter NCC.
Sci Adv, 8, 2022
|
|
7S8D
 
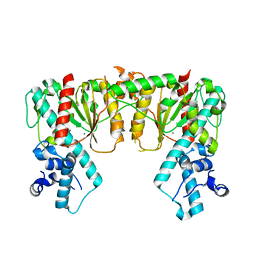 | | Structure of DNA-free SgrAI | | Descriptor: | CALCIUM ION, SgraIR restriction enzyme | | Authors: | Horton, N.C. | | Deposit date: | 2021-09-17 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Pretransition state and apo structures of the filament-forming enzyme SgrAI elucidate mechanisms of activation and substrate specificity.
J.Biol.Chem., 298, 2022
|
|
8FND
 
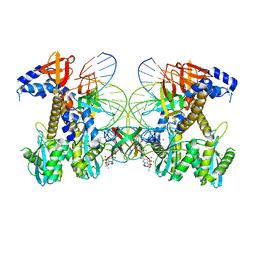 | | Structure of E138K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNP
 
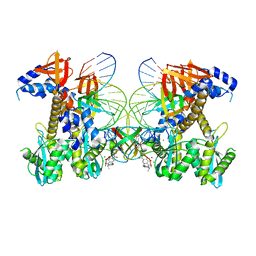 | | Structure of E138K/G140S/Q148H HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNM
 
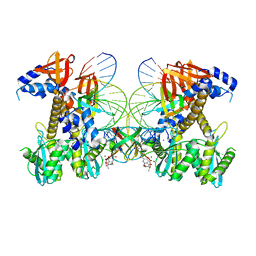 | | Structure of G140A/Q148K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
