4CT3
 
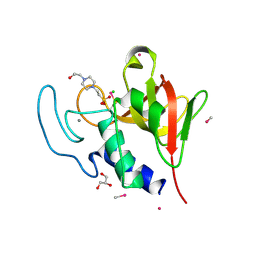 | | Methylmercury chloride derivative structure of the lytic CHAPK domain of the endolysin LysK from Staphylococcus aureus bacteriophage K | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sanz-Gaitero, M, Keary, R, Garcia-Doval, C, Coffey, A, van Raaij, M.J. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of the lytic CHAP(K) domain of the endolysin LysK from Staphylococcus aureus bacteriophage K.
Virol. J., 11, 2014
|
|
4CSH
 
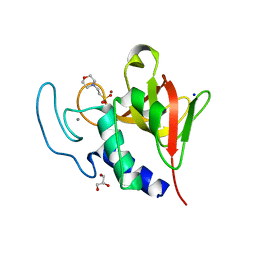 | | Native structure of the lytic CHAPK domain of the endolysin LysK from Staphylococcus aureus bacteriophage K | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Sanz-Gaitero, M, Keary, R, Garcia-Doval, C, Coffey, A, van Raaij, M.J. | | Deposit date: | 2014-03-07 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the lytic CHAP(K) domain of the endolysin LysK from Staphylococcus aureus bacteriophage K.
Virol. J., 11, 2014
|
|
8P8E
 
 | |
6SFE
 
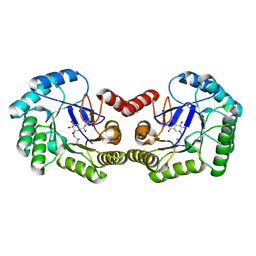 | | CRYSTAL STRUCTURE OF DHQ1 FROM SALMONELLA TYPHI COVALENTLY MODIFIED BY COMPOUND 7 | | Descriptor: | (1~{S},3~{S},4~{S},5~{R})-3-(aminomethyl)-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Lence, E, Maneiro, M, Thompson, R, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Self-Immolation of a Bacterial Dehydratase Enzyme by its Epoxide Product.
Chemistry, 26, 2020
|
|
6SFH
 
 | | CRYSTAL STRUCTURE OF DHQ1 FROM Staphylococcus aureus COVALENTLY MODIFIED BY LIGAND 7 | | Descriptor: | (1~{S},3~{S},4~{S},5~{R})-3-(aminomethyl)-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase, LITHIUM ION, ... | | Authors: | Sanz-Gaitero, M, Lence, E, Maneiro, M, Thompson, R, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-15 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Self-Immolation of a Bacterial Dehydratase Enzyme by its Epoxide Product.
Chemistry, 26, 2020
|
|
6SFG
 
 | | CRYSTAL STRUCTURE OF DHQ1 FROM SALMONELLA TYPHI COVALENTLY MODIFIED BY COMPOUND 9 | | Descriptor: | (1~{R},3~{S},4~{R},5~{R})-3-methyl-4,5-bis(hydroxyl)cyclohexane-1-carboxylic acid, (1~{S},3~{R},4~{S},5~{R})-3-methyl-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic Acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Lence, E, Maneiro, M, Thompson, R, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Self-Immolation of a Bacterial Dehydratase Enzyme by its Epoxide Product.
Chemistry, 26, 2020
|
|
6H5C
 
 | | Crystal structure of DHQ1 from Salmonella typhi covalently modified by ligand 1 | | Descriptor: | (1~{S},3~{R},4~{S},5~{R})-3-methyl-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic Acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Maneiro, M, Lence, E, Otero, J.M, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Hydroxylammonium Derivatives for Selective Active-site Lysine Modification in the Anti-virulence Bacterial Target DHQ1 Enzyme.
Org Chem Front, 9, 2019
|
|
6H5D
 
 | | Crystal structure of DHQ1 from Salmonella typhi covalently modified by ligand 2 | | Descriptor: | (1~{S},3~{S},4~{R},5~{R})-3-methyl-1,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Maneiro, M, Lence, E, Otero, J.M, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hydroxylammonium Derivatives for Selective Active-site Lysine Modification in the Anti-virulence Bacterial Target DHQ1 Enzyme.
Org Chem Front, 9, 2019
|
|
6H5G
 
 | | Crystal structure of DHQ1 from Salmonella typhi covalently modified by ligand 3 | | Descriptor: | (1~{R},3~{S},4~{R},5~{R})-3-methyl-4,5-bis(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Maneiro, M, Lence, E, Otero, J.M, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Hydroxylammonium Derivatives for Selective Active-site Lysine Modification in the Anti-virulence Bacterial Target DHQ1 Enzyme.
Org Chem Front, 6, 2019
|
|
6H5J
 
 | | Crystal structure of DHQ1 from Salmonella typhi covalently modified by ligand 4 | | Descriptor: | (3~{R})-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Otero, J.M, Maneiro, M, Lence, E, Sanz-Gaitero, M, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Hydroxylammonium Derivatives for Selective Active-site Lysine Modification in the Anti-virulence Bacterial Target DHQ1 Enzyme.
Org Chem Front, 6, 2019
|
|
4UE0
 
 | |
8B2C
 
 | | Crystal structure of type I dehydroquinase from Salmonella typhi inhibited by an epoxide derivative | | Descriptor: | (1~{S},2~{R},4~{R},5~{S},6~{S})-2,4,5-trihydroxy-7-oxabicyclo[4.1.0]heptane-2-carboxylic acid, 3-dehydroquinate dehydratase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Otero, J.M, Rodriguez, A, Maneiro, M, Lence, E, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2022-09-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Quinate-based ligands for irreversible inactivation of the bacterial virulence factor DHQ1 enzyme-A molecular insight.
Front Mol Biosci, 10, 2023
|
|
8B2A
 
 | | Crystal structure of type I dehydroquinase from Salmonella typhi inhibited by an epoxide derivative | | Descriptor: | (4R,5R)-3-amino-4,5-dihydroxy-cyclohexene-1-carboxylic acid, 3-dehydroquinate dehydratase, CHLORIDE ION, ... | | Authors: | Otero, J.M, Rodriguez, A, Maneiro, M, Lence, E, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2022-09-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Quinate-based ligands for irreversible inactivation of the bacterial virulence factor DHQ1 enzyme-A molecular insight.
Front Mol Biosci, 10, 2023
|
|
8B2B
 
 | | Crystal structure of type I dehydroquinase from Salmonella typhi inhibited by an epoxide derivative | | Descriptor: | (4R,5R)-3-amino-4,5-dihydroxy-cyclohexene-1-carboxylic acid, 3-dehydroquinate dehydratase, SODIUM ION | | Authors: | Otero, J.M, Rodriguez, A, Maneiro, M, Lence, E, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2022-09-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Quinate-based ligands for irreversible inactivation of the bacterial virulence factor DHQ1 enzyme-A molecular insight.
Front Mol Biosci, 10, 2023
|
|
6ZJA
 
 | | Helicobacter pylori urease with inhibitor bound in the active site | | Descriptor: | 2-{[1-(3,5-dimethylphenyl)-1H-imidazol-2-yl]sulfanyl}-N-hydroxyacetamide, NICKEL (II) ION, Urease subunit alpha, ... | | Authors: | Luecke, H, Cunha, E. | | Deposit date: | 2020-06-28 | | Release date: | 2020-12-23 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Cryo-EM structure of Helicobacter pylori urease with an inhibitor in the active site at 2.0 angstrom resolution.
Nat Commun, 12, 2021
|
|
6QSU
 
 | | Helicobacter pylori urease with BME bound in the active site | | Descriptor: | BETA-MERCAPTOETHANOL, NICKEL (II) ION, Urease subunit alpha, ... | | Authors: | Luecke, H, Cunha, E. | | Deposit date: | 2019-02-22 | | Release date: | 2021-01-20 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM structure of Helicobacter pylori urease with an inhibitor in the active site at 2.0 angstrom resolution.
Nat Commun, 12, 2021
|
|
