2GC8
 
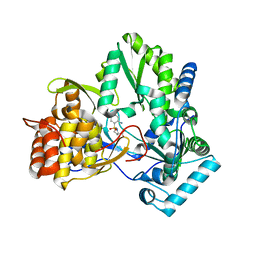 | | Structure of a Proline Sulfonamide Inhibitor Bound to HCV NS5b Polymerase | | Descriptor: | 1-[(2-AMINO-4-CHLORO-5-METHYLPHENYL)SULFONYL]-L-PROLINE, RNA-directed RNA polymerase | | Authors: | Gopalsamy, A, Chopra, R, Lim, K, Ciszewski, G, Shi, M, Curran, K.J, Sukits, S.F, Svenson, K, Bard, J, Ellingboe, J.W, Agarwal, A, Krishnamurthy, G, Howe, A.Y, Orlowski, M, Feld, B, O'connell, J, Mansour, T.S. | | Deposit date: | 2006-03-13 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Proline Sulfonamides as Potent and Selective Hepatitis C Virus NS5b Polymerase Inhibitors. Evidence for a New NS5b Polymerase Binding Site.
J.Med.Chem., 49, 2006
|
|
1LDA
 
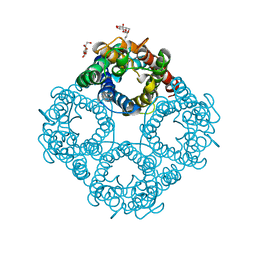 | | CRYSTAL STRUCTURE OF THE E. COLI GLYCEROL FACILITATOR (GLPF) WITHOUT SUBSTRATE GLYCEROL | | Descriptor: | Glycerol uptake facilitator protein, octyl beta-D-glucopyranoside | | Authors: | Nollert, P, Miercke, L.J.W, O'Connell, J, Stroud, R.M. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Control of the selectivity of the aquaporin water channel family by global orientational tuning.
Science, 296, 2002
|
|
1LDI
 
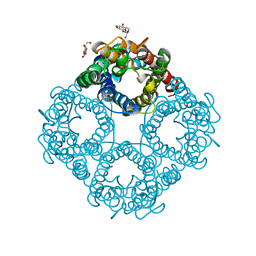 | | CRYSTAL STRUCTURE OF THE E. COLI GLYCEROL FACILITATOR (GLPF) WITHOUT SUBSTRATE GLYCEROL | | Descriptor: | Glycerol uptake facilitator protein, octyl beta-D-glucopyranoside | | Authors: | Nollert, P, Miercke, L.J.W, O'Connell, J, Stroud, R.M. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Control of the selectivity of the aquaporin water channel family by global orientational tuning.
Science, 296, 2002
|
|
1LDF
 
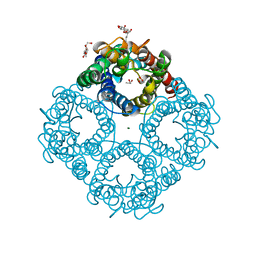 | | CRYSTAL STRUCTURE OF THE E. COLI GLYCEROL FACILITATOR (GLPF) MUTATION W48F, F200T | | Descriptor: | GLYCEROL, Glycerol uptake facilitator protein, MAGNESIUM ION, ... | | Authors: | Nollert, P, Miercke, L.J.W, O'Connell, J, Stroud, R.M. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Control of the selectivity of the aquaporin water channel family by global orientational tuning.
Science, 296, 2002
|
|
1MZJ
 
 | | Crystal Structure of the Priming beta-Ketosynthase from the R1128 Polyketide Biosynthetic Pathway | | Descriptor: | ACETYL GROUP, Beta-ketoacylsynthase III, COENZYME A | | Authors: | Pan, H, Tsai, S.C, Meadows, E.S, Miercke, L.J.W, Keatinge-Clay, A, O'Connell, J, Khosla, C, Stroud, R.M. | | Deposit date: | 2002-10-08 | | Release date: | 2002-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the priming beta-ketosynthase from the
R1128 polyketide biosynthetic pathway
Structure, 10
|
|
1RD4
 
 | | An allosteric inhibitor of LFA-1 bound to its I-domain | | Descriptor: | 1-ACETYL-4-(4-{4-[(2-ETHOXYPHENYL)THIO]-3-NITROPHENYL}PYRIDIN-2-YL)PIPERAZINE, Integrin alpha-L | | Authors: | Crump, M.P, Ceska, T.A, Spyracopoulos, L, Henry, A, Archibald, S.C, Alexander, R, Taylor, R.J, Findlow, S.C, O'Connell, J, Robinson, M.K, Shock, A. | | Deposit date: | 2003-11-05 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of an allosteric inhibitor of LFA-1 bound to the I-domain studied by crystallography, NMR, and calorimetry
Biochemistry, 43, 2004
|
|
2CCX
 
 | |
6OOZ
 
 | | Asymmetric hTNF-alpha | | Descriptor: | (S)-{1-[(2,5-dimethylphenyl)methyl]-1H-benzimidazol-2-yl}(pyridin-4-yl)methanol, CHLORIDE ION, Tumor necrosis factor | | Authors: | Arakaki, T.L, Edwards, T.E, Fox III, D, Lecomte, F, Ceska, T. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Small molecules that inhibit TNF signalling by stabilising an asymmetric form of the trimer.
Nat Commun, 10, 2019
|
|
6OP0
 
 | | Asymmetric hTNF-alpha | | Descriptor: | (R)-{1-[(2,5-dimethylphenyl)methyl]-6-(1-methyl-1H-pyrazol-4-yl)-1H-benzimidazol-2-yl}(pyridin-4-yl)methanol, Tumor necrosis factor | | Authors: | Arakaki, T.L, Edwards, T.E, Fairman, J.W, Davies, D.R, Foley, A, Ceska, T. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Small molecules that inhibit TNF signalling by stabilising an asymmetric form of the trimer.
Nat Commun, 10, 2019
|
|
6OOY
 
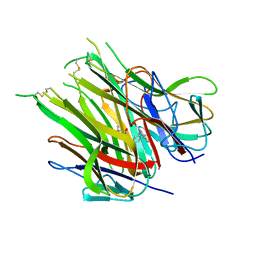 | | Asymmetric hTNF-alpha | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Tumor necrosis factor, {1-[(2,5-dimethylphenyl)methyl]-1H-benzimidazol-2-yl}methanol | | Authors: | Arakaki, T.L, Edwards, T.E, Ceska, T. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Small molecules that inhibit TNF signalling by stabilising an asymmetric form of the trimer.
Nat Commun, 10, 2019
|
|
1HRI
 
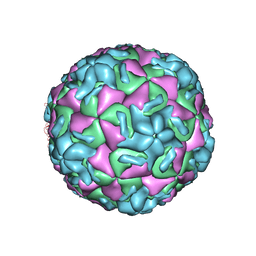 | | STRUCTURE DETERMINATION OF ANTIVIRAL COMPOUND SCH 38057 COMPLEXED WITH HUMAN RHINOVIRUS 14 | | Descriptor: | 1-[6-(2-CHLORO-4-METHYXYPHENOXY)-HEXYL]-IMIDAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Zhang, A, Nanni, R.G, Oren, D.A, Arnold, E. | | Deposit date: | 1992-10-01 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure determination of antiviral compound SCH 38057 complexed with human rhinovirus 14.
J.Mol.Biol., 230, 1993
|
|
2QE5
 
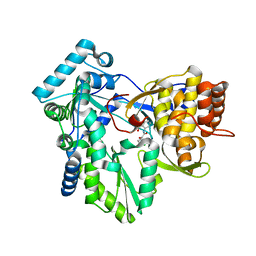 | |
2QE2
 
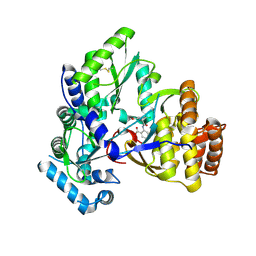 | | Structure of HCV NS5B Bound to an Anthranilic Acid Inhibitor | | Descriptor: | 2-{[N-(2-ACETYL-5-CHLORO-4-FLUOROPHENYL)GLYCYL]AMINO}BENZOIC ACID, RNA-directed RNA polymerase | | Authors: | Chopra, R, Svenson, K, Bard, J. | | Deposit date: | 2007-06-22 | | Release date: | 2007-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of Anthranilic Acid Derivatives as a Novel Class of Allosteric Inhibitors of Hepatitis C NS5B Polymerase
J.Med.Chem., 50, 2007
|
|
1EAH
 
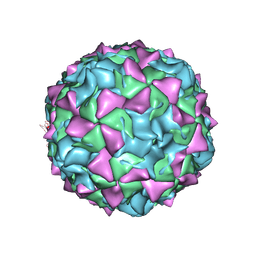 | | PV2L COMPLEXED WITH ANTIVIRAL AGENT SCH48973 | | Descriptor: | 1[2-CHLORO-4-METHOXY-PHENYL-OXYMETHYL]-4-[2,6-DICHLORO-PHENYL-OXYMETHYL]-BENZENE, MYRISTIC ACID, POLIOVIRUS TYPE 2 COAT PROTEINS VP1 TO VP4 | | Authors: | Lentz, K, Arnold, E. | | Deposit date: | 1997-07-22 | | Release date: | 1998-09-16 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of poliovirus type 2 Lansing complexed with antiviral agent SCH48973: comparison of the structural and biological properties of three poliovirus serotypes.
Structure, 5, 1997
|
|
8DIC
 
 | |
8DIB
 
 | |
8DIG
 
 | |
8DII
 
 | |
8DIE
 
 | |
8DIF
 
 | |
8DIH
 
 | | Virtual screening for novel SARS-CoV-2 main protease non-covalent and covalent inhibitors | | Descriptor: | (1P,1'R)-1-(isoquinolin-4-yl)-2',3'-dihydrospiro[imidazolidine-4,1'-indene]-2,5-dione, 3C-like proteinase nsp5 | | Authors: | Singh, I, Shoichet, B.K. | | Deposit date: | 2022-06-29 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Large library docking for novel SARS-CoV-2 main protease non-covalent and covalent inhibitors.
Protein Sci., 32, 2023
|
|
8DID
 
 | |
3C02
 
 | |
7KP9
 
 | | asymmetric hTNF-alpha | | Descriptor: | 1-{[2-(difluoromethoxy)phenyl]methyl}-2-methyl-6-[6-(piperazin-1-yl)pyridin-3-yl]-1H-benzimidazole, Tumor necrosis factor | | Authors: | Arakaki, T.L, Abendroth, J, Fairman, J.W, Foley, A, Ceska, T. | | Deposit date: | 2020-11-10 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into the disruption of TNF-TNFR1 signalling by small molecules stabilising a distorted TNF.
Nat Commun, 12, 2021
|
|
7KP7
 
 | | asymmetric mTNF-alpha hTNFR1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Tumor necrosis factor, ... | | Authors: | Arakaki, T.L, Fox III, D, Edwards, T.E, Foley, A, Ceska, T. | | Deposit date: | 2020-11-10 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into the disruption of TNF-TNFR1 signalling by small molecules stabilising a distorted TNF.
Nat Commun, 12, 2021
|
|
