8Q96
 
 | | P301S Tau Filaments from the Brains of Tg2541 Transgenic Mouse Line | | Descriptor: | Isoform Tau-E of Microtubule-associated protein tau, Unknown protein | | Authors: | Schweighauser, M, Murzin, A.G, Macdonald, J, Lavenir, I, Crowther, R.A, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-19 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryo-EM structures of tau filaments from the brains of mice transgenic for human mutant P301S Tau.
Acta Neuropathol Commun, 11, 2023
|
|
8Q92
 
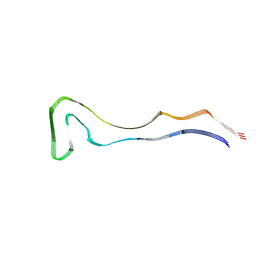 | | P301S Tau Filaments from the Brains of PS19 Transgenic Mouse Line | | Descriptor: | Microtubule-associated protein tau | | Authors: | Schweighauser, M, Murzin, A.G, Macdonald, J, Lavenir, I, Crowther, R.A, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-19 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cryo-EM structures of tau filaments from the brains of mice transgenic for human mutant P301S Tau.
Acta Neuropathol Commun, 11, 2023
|
|
7Q4M
 
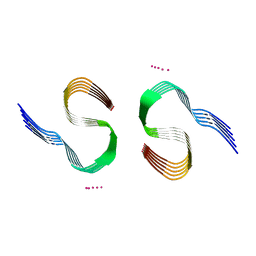 | | Type II beta-amyloid 42 Filaments from Human Brain | | Descriptor: | Amyloid-beta precursor protein, UNKNOWN ATOM OR ION | | Authors: | Yang, Y, Arseni, D, Zhang, W, Huang, M, Lovestam, S.K.A, Schweighauser, M, Kotecha, A, Murzin, A.G, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Garringer, H.J, Gelpi, E, Newell, K.L, Kovacs, G.G, Vidal, R, Ghetti, B, Falcon, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-11-01 | | Release date: | 2021-11-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of amyloid-beta 42 filaments from human brains.
Science, 375, 2022
|
|
7Q4B
 
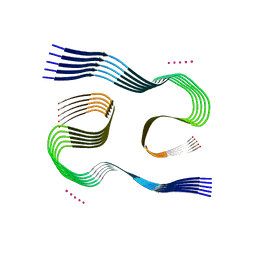 | | Type I beta-amyloid 42 Filaments from Human Brain | | Descriptor: | Amyloid-beta precursor protein, UNKNOWN ATOM OR ION | | Authors: | Yang, Y, Arseni, D, Zhang, W, Huang, M, Lovestam, S.K.A, Schweighauser, M, Kotecha, A, Murzin, A.G, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Garringer, H.J, Gelpi, E, Newell, K.L, Kovacs, G.G, Vidal, R, Ghetti, B, Falcon, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-10-30 | | Release date: | 2021-11-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structures of amyloid-beta 42 filaments from human brains.
Science, 375, 2022
|
|
8BG9
 
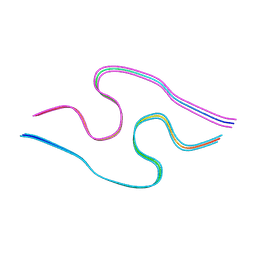 | | Murine amyloid-beta filaments with the Arctic mutation (E22G) from APP(NL-G-F) mouse brains | ABeta | | Descriptor: | Amyloid-beta protein 40 | | Authors: | Yang, Y, Zhang, W.J, Murzin, A.G, Schweighauser, M, Huang, M, Lovestam, S.K.A, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Ghetti, B, Graff, C, Kumar, A, Nordber, A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2022-10-27 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of amyloid-beta filaments with the Arctic mutation (E22G) from human and mouse brains.
Acta Neuropathol, 145, 2023
|
|
8BG0
 
 | | Amyloid-beta tetrameric filaments with the Arctic mutation (E22G) from Alzheimer's disease brains | ABeta40 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Yang, Y, Zhang, W.J, Murzin, A.G, Schweighauser, M, Huang, M, Lovestam, S.K.A, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Ghetti, B, Graff, C, Kumar, A, Nordber, A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2022-10-27 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (1.99 Å) | | Cite: | Cryo-EM structures of amyloid-beta filaments with the Arctic mutation (E22G) from human and mouse brains.
Acta Neuropathol, 145, 2023
|
|
8BFZ
 
 | | Amyloid-beta 42 filaments extracted from the human brain with Arctic mutation (E22G) of Alzheimer's disease | ABeta42 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Yang, Y, Zhang, W.J, Murzin, A.G, Schweighauser, M, Huang, M, Lovestam, S.K.A, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Ghetti, B, Graff, C, Kumar, A, Nordberg, A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2022-10-27 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of amyloid-beta filaments with the Arctic mutation (E22G) from human and mouse brains.
Acta Neuropathol, 145, 2023
|
|
1UZ1
 
 | | Family 1 b-glucosidase from Thermotoga maritima in complex with isofagomine lactam | | Descriptor: | (3S,4R,5R)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)PIPERIDIN-2-ONE, BETA-GLUCOSIDASE A | | Authors: | Gloster, T.M, Macdonald, J, Stick, R.V, Davies, G.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Common Inhibition of Both -Glucosidases and -Mannosidases by Isofagomine Lactam Reflects Different Conformational Itineraries for Pyranoside Hydrolysis
Chembiochem, 5, 2004
|
|
1OCN
 
 | | Mutant D416A of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with a cellobio-derived isofagomine at 1.3 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, CALCIUM ION, ... | | Authors: | Varrot, A, Macdonald, J, Stick, R.V, Pell, G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2003-02-09 | | Release date: | 2003-05-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Distortion of a Cellobio-Derived Isofagomine Highlights the Potential Conformational Itinerary of Inverting Beta-Glucosidases
Chem.Commun.(Camb.), 21, 2003
|
|
1OCQ
 
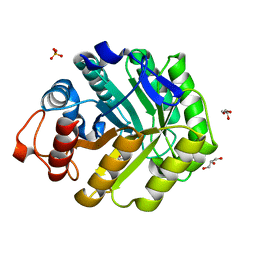 | | COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.08 ANGSTROM RESOLUTION with cellobio-derived isofagomine | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ENDOGLUCANASE 5A, GLYCEROL, ... | | Authors: | Varrot, A, Macdonald, J, Stick, R.V, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-02-09 | | Release date: | 2003-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Direct Observation of the Protonation State of an Imino Sugar Glycosidase Inhibitor Upon Binding
J.Am.Chem.Soc., 125, 2003
|
|
5HCU
 
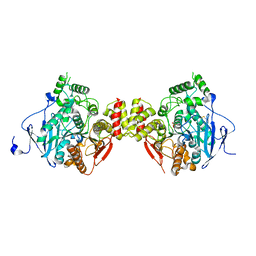 | |
1UZ4
 
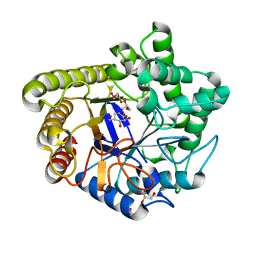 | | Common inhibition of beta-glucosidase and beta-mannosidase by isofagomine lactam reflects different conformational intineraries for glucoside and mannoside hydrolysis | | Descriptor: | (3S,4R,5R)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)PIPERIDIN-2-ONE, GLYCEROL, MAN5A, ... | | Authors: | Vincent, F, Davies, G.J. | | Deposit date: | 2004-03-04 | | Release date: | 2004-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Common Inhibition of Both Beta-Glucosidases and Beta-Mannosidases by Isofagomine Lactam Reflects Different Conformational Itineraries for Pyranoside Hydrolysis
Chembiochem, 5, 2004
|
|
3I2G
 
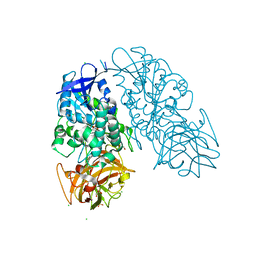 | | Cocaine Esterase with mutation G173Q, bound to DTT adduct | | Descriptor: | (4S,5S)-4,5-BIS(MERCAPTOMETHYL)-1,3-DIOXOLAN-2-OL, CHLORIDE ION, Cocaine esterase, ... | | Authors: | Tesmer, J.J.G, Nance, M.R. | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of thermostabilizing mutations of cocaine esterase.
Protein Eng.Des.Sel., 23, 2010
|
|
3I2K
 
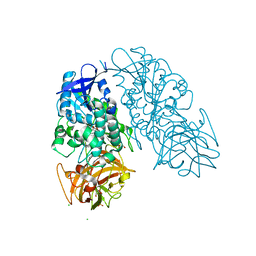 | | Cocaine Esterase, wild type, bound to a DTT adduct | | Descriptor: | (4S,5S)-4,5-BIS(MERCAPTOMETHYL)-1,3-DIOXOLAN-2-OL, CHLORIDE ION, Cocaine esterase, ... | | Authors: | Tesmer, J.J.G, Nance, M.R. | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural analysis of thermostabilizing mutations of cocaine esterase.
Protein Eng.Des.Sel., 23, 2010
|
|
3I2H
 
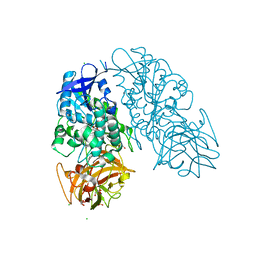 | | Cocaine Esterase with mutation L169K, bound to DTT adduct | | Descriptor: | (4S,5S)-4,5-BIS(MERCAPTOMETHYL)-1,3-DIOXOLAN-2-OL, CHLORIDE ION, Cocaine esterase, ... | | Authors: | Tesmer, J.J.G, Nance, M.R. | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural analysis of thermostabilizing mutations of cocaine esterase.
Protein Eng.Des.Sel., 23, 2010
|
|
3I2J
 
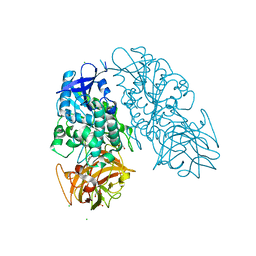 | | Cocaine Esterase, wild type, without a ligand | | Descriptor: | CHLORIDE ION, Cocaine esterase, GLYCEROL, ... | | Authors: | Tesmer, J.J.G, Nance, M.R. | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural analysis of thermostabilizing mutations of cocaine esterase.
Protein Eng.Des.Sel., 23, 2010
|
|
3I2I
 
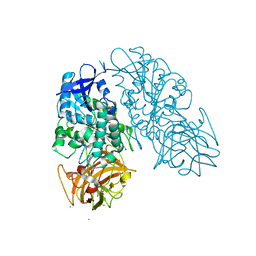 | | Cocaine Esterase with mutation T172R, bound to DTT adduct | | Descriptor: | (4S,5S)-4,5-BIS(MERCAPTOMETHYL)-1,3-DIOXOLAN-2-OL, CHLORIDE ION, Cocaine esterase, ... | | Authors: | Tesmer, J.J.G, Nance, M.R. | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural analysis of thermostabilizing mutations of cocaine esterase.
Protein Eng.Des.Sel., 23, 2010
|
|
3I2F
 
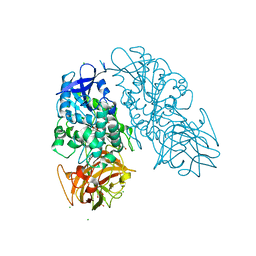 | | Cocaine Esterase with mutations T172R / G173Q, bound to DTT adduct | | Descriptor: | (4S,5S)-4,5-BIS(MERCAPTOMETHYL)-1,3-DIOXOLAN-2-OL, CHLORIDE ION, Cocaine esterase, ... | | Authors: | Tesmer, J.J.G, Nance, M.R. | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of thermostabilizing mutations of cocaine esterase.
Protein Eng.Des.Sel., 23, 2010
|
|
5DTJ
 
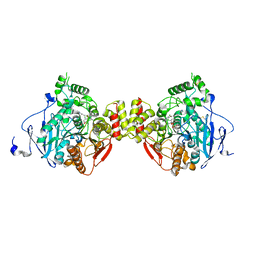 | |
5DTI
 
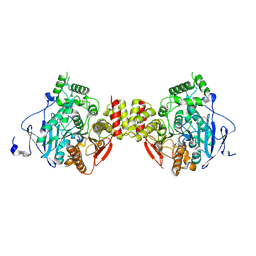 | | Crystal structure of mouse acetylcholinesterase | | Descriptor: | Acetylcholinesterase | | Authors: | Tran, T.H, Tong, L. | | Deposit date: | 2015-09-18 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Discovery of New Classes of Compounds that Reactivate Acetylcholinesterase Inhibited by Organophosphates.
Chembiochem, 16, 2015
|
|
