1IQO
 
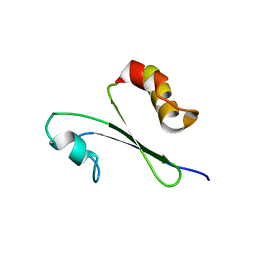 | | Solution structure of MTH1880 from methanobacterium thermoautotrophicum | | Descriptor: | HYPOTHETICAL PROTEIN MTH1880 | | Authors: | Lee, C.H, Shin, J, Bang, E, Jung, J.W, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2001-07-23 | | Release date: | 2002-07-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel calcium binding protein, MTH1880, from Methanobacterium thermoautotrophicum.
Protein Sci., 13, 2004
|
|
1IQS
 
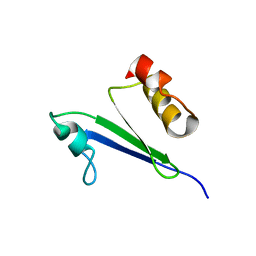 | | Minimized average structure of MTH1880 from Methanobacterium Thermoautotrophicum | | Descriptor: | MTH1880 | | Authors: | Lee, C.H, Shin, J, Bang, E, Jung, J.W, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2001-07-29 | | Release date: | 2002-07-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel calcium binding protein, MTH1880, from Methanobacterium thermoautotrophicum.
Protein Sci., 13, 2004
|
|
8IDU
 
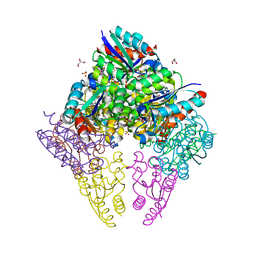 | | Crystal structure of substrate bound-form dehydroquinate dehydratase from Corynebacterium glutamicum | | Descriptor: | 1,3,4-TRIHYDROXY-5-OXO-CYCLOHEXANECARBOXYLIC ACID, 3-dehydroquinate dehydratase, GLYCEROL, ... | | Authors: | Lee, C.H, Kim, S, Kim, K.-J. | | Deposit date: | 2023-02-14 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Analysis of 3-Dehydroquinate Dehydratase from Corynebacterium glutamicum .
J Microbiol Biotechnol., 33, 2023
|
|
8IDR
 
 | |
6CNN
 
 | |
6CNO
 
 | |
6CNM
 
 | | Cryo-EM structure of the human SK4/calmodulin channel complex | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Calmodulin-1, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Lee, C.H, MacKinnon, R. | | Deposit date: | 2018-03-08 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Activation mechanism of a human SK-calmodulin channel complex elucidated by cryo-EM structures.
Science, 360, 2018
|
|
3TNU
 
 | | Heterocomplex of coil 2B domains of human intermediate filament proteins, keratin 5 (KRT5) and keratin 14 (KRT14) | | Descriptor: | Keratin, type I cytoskeletal 14, type II cytoskeletal 5 | | Authors: | Lee, C.H, Kim, M.S, Leahy, D.J, Coulombe, P.A. | | Deposit date: | 2011-09-02 | | Release date: | 2012-06-20 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structural basis for heteromeric assembly and perinuclear organization of keratin filaments.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6JFV
 
 | | The crystal structure of 2B-2B complex from keratins 5 and 14 (C367A mutant of K14) | | Descriptor: | Keratin, type I cytoskeletal 14, type II cytoskeletal 5 | | Authors: | Kim, M.S, Lee, C.H, Coulombe, P.A, Leahy, D.J. | | Deposit date: | 2019-02-12 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Function Analyses of a Keratin Heterotypic Complex Identify Specific Keratin Regions Involved in Intermediate Filament Assembly.
Structure, 28, 2020
|
|
8ZUG
 
 | |
1RYJ
 
 | | Solution NMR Structure of Protein Mth1743 from Methanobacterium thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH1743_1_70; Northeast Structural Genomics Consortium Target TT526. | | Descriptor: | unknown | | Authors: | Yee, A, Chang, X, Pineda-Lucena, A, Wu, B, Semesi, A, Le, B, Ramelot, T, Lee, G.M, Bhattacharyya, S, Gutierrez, P, Denisov, A, Lee, C.H, Cort, J.R, Kozlov, G, Liao, J, Finak, G, Chen, L, Wishart, D, Lee, W, McIntosh, L.P, Gehring, K, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | AN NMR APPROACH TO STRUCTURAL PROTEOMICS
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
9B1H
 
 | | Human urate transporter 1 URAT1 in complex with lesinurad | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 22 member 12, lesinurad | | Authors: | Dai, Y, Lee, C.H. | | Deposit date: | 2024-03-13 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Transport mechanism and structural pharmacology of human urate transporter URAT1.
Cell Res., 2024
|
|
9B1K
 
 | | Urate bound human URAT1 in the occluded state | | Descriptor: | Solute carrier family 22 member 12, URIC ACID | | Authors: | Dai, Y, Lee, C.H. | | Deposit date: | 2024-03-13 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Transport mechanism and structural pharmacology of human urate transporter URAT1.
Cell Res., 2024
|
|
9B1O
 
 | |
9B1J
 
 | | Urate bound human URAT1 in the inward-facing state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, Solute carrier family 22 member 12, ... | | Authors: | Dai, Y, Lee, C.H. | | Deposit date: | 2024-03-13 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Transport mechanism and structural pharmacology of human urate transporter URAT1.
Cell Res., 2024
|
|
9B1G
 
 | | Human urate transporter 1 URAT1 in complex with dotinurad | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 22 member 12, dotinurad | | Authors: | Dai, Y, Lee, C.H. | | Deposit date: | 2024-03-13 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Transport mechanism and structural pharmacology of human urate transporter URAT1.
Cell Res., 2024
|
|
9B1L
 
 | | Urate bound human URAT1 in the outward-facing state. | | Descriptor: | Solute carrier family 22 member 12, URIC ACID | | Authors: | Dai, Y, Lee, C.H. | | Deposit date: | 2024-03-13 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Transport mechanism and structural pharmacology of human urate transporter URAT1.
Cell Res., 2024
|
|
9B1N
 
 | |
9B1F
 
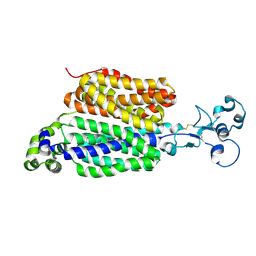 | | Human urate transporter 1 URAT1 in apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 22 member 12 | | Authors: | Dai, Y, Lee, C.H. | | Deposit date: | 2024-03-13 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Transport mechanism and structural pharmacology of human urate transporter URAT1.
Cell Res., 2024
|
|
9B1I
 
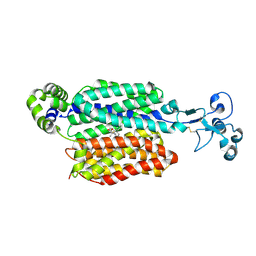 | | Human urate transporter 1 URAT1 in complex with verinurad | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 22 member 12, verinurad | | Authors: | Dai, Y, Lee, C.H. | | Deposit date: | 2024-03-13 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Transport mechanism and structural pharmacology of human urate transporter URAT1.
Cell Res., 2024
|
|
9B1M
 
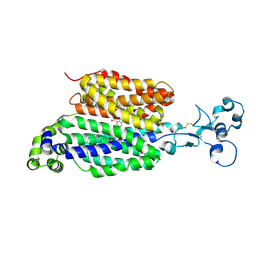 | | Pyrazinoate bound human URAT1 in the inward-facing state (site1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PYRAZINE-2-CARBOXYLIC ACID, Solute carrier family 22 member 12 | | Authors: | Dai, Y, Lee, C.H. | | Deposit date: | 2024-03-13 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Transport mechanism and structural pharmacology of human urate transporter URAT1.
Cell Res., 2024
|
|
2BR6
 
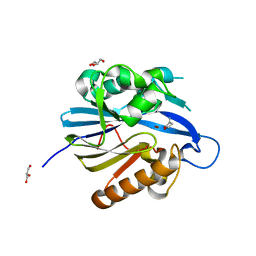 | | Crystal Structure of Quorum-Quenching N-Acyl Homoserine Lactone Lactonase | | Descriptor: | AIIA-LIKE PROTEIN, GLYCEROL, HOMOSERINE LACTONE, ... | | Authors: | Kim, M.H, Choi, W.C, Kang, H.O, Kang, B.S, Kim, K.J, Derewenda, Z.S, Lee, J.K, Oh, T.K, Lee, C.H. | | Deposit date: | 2005-05-03 | | Release date: | 2005-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Molecular Structure and Catalytic Mechanism of a Quorum-Quenching N-Acyl-L-Homoserine Lactone Hydrolase.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BTN
 
 | | Crystal Structure and Catalytic Mechanism of the Quorum-Quenching N- Acyl Homoserine Lactone Hydrolase | | Descriptor: | AIIA-LIKE PROTEIN, GLYCEROL, ZINC ION | | Authors: | Kim, M.H, Choi, W.C, Kang, H.O, Lee, J.S, Kang, B.S, Kim, K.J, Derewenda, Z.S, Oh, T.K, Lee, C.H, Lee, J.K. | | Deposit date: | 2005-06-03 | | Release date: | 2005-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Molecular Structure and Catalytic Mechanism of a Quorum-Quenching N-Acyl-L-Homoserine Lactone Hydrolase.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
8T6B
 
 | | Human VMAT2 in complex with serotonin | | Descriptor: | SEROTONIN, Synaptic vesicular amine transporter | | Authors: | Pidathala, S, Dai, Y, Lee, C.H. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Mechanisms of neurotransmitter transport and drug inhibition in human VMAT2.
Nature, 623, 2023
|
|
8T69
 
 | | Human VMAT2 in complex with tetrabenazine | | Descriptor: | Synaptic vesicular amine transporter, tetrabenazine | | Authors: | Pidathala, S, Dai, Y, Lee, C.H. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Mechanisms of neurotransmitter transport and drug inhibition in human VMAT2.
Nature, 623, 2023
|
|
