6L33
 
 | |
4WYS
 
 | | Crystal structure of thiolase from Escherichia coli | | Descriptor: | Acetyl-CoA acetyltransferase | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2014-11-18 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4WYR
 
 | | Crystal structure of thiolase mutation (V77Q,N153Y,A286K) from Clostridium acetobutylicum | | Descriptor: | Acetyl-CoA acetyltransferase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2014-11-18 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4XL4
 
 | | Crystal structure of thiolase from Clostridium acetobutylicum in complex with CoA | | Descriptor: | Acetyl-CoA acetyltransferase, COENZYME A, GLYCEROL | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2015-01-13 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4XL2
 
 | | Crystal structure of oxidized form of thiolase from Clostridium acetobutylicum | | Descriptor: | ACETATE ION, Acetyl-CoA acetyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2015-01-13 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4XL3
 
 | | Crystal structure of reduced form of thiolase from Clostridium acetobutylicum | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2015-01-13 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4XGS
 
 | |
7VG4
 
 | | 10,5-methenyltetrahydrofolate cyclohydrolase from Methylobacterium extorquens AM1 strain | | Descriptor: | Methenyltetrahydrofolate cyclohydrolase | | Authors: | Kim, S, Lee, S, Kim, I.-K, Seo, H, Kim, K.-J. | | Deposit date: | 2021-09-14 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural insight into a molecular mechanism of methenyltetrahydrofolate cyclohydrolase from Methylobacterium extorquens AM1.
Int.J.Biol.Macromol., 202, 2022
|
|
7YA3
 
 | |
7YA4
 
 | |
7VG5
 
 | | 10,5-methenyltetrahydrofolate cyclohydrolase from Methylobacterium extorquens AM1 with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, Methenyltetrahydrofolate cyclohydrolase | | Authors: | Kim, S, Lee, S, Kim, I.-K, Seo, H, Kim, K.-J. | | Deposit date: | 2021-09-14 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insight into a molecular mechanism of methenyltetrahydrofolate cyclohydrolase from Methylobacterium extorquens AM1.
Int.J.Biol.Macromol., 202, 2022
|
|
4ZTT
 
 | | Crystal structures of ferritin mutants reveal diferric-peroxo intermediates | | Descriptor: | FE (II) ION, FE (III) ION, GLYCEROL, ... | | Authors: | Kim, S, Park, Y.H, Jung, S.W, Seok, J.H, Chung, Y.B, Lee, D.B, Gowda, G, Lee, J.H, Han, H.R, Cho, A.E, Lee, C, Chung, M.S, Kim, K.H. | | Deposit date: | 2015-05-15 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis of Novel Iron-Uptake Route and Reaction Intermediates in Ferritins from Gram-Negative Bacteria.
J. Mol. Biol., 428, 2016
|
|
7DNZ
 
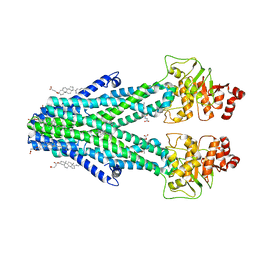 | | Cryo-EM structure of the human ABCB6 (Hemin and GSH-bound) | | Descriptor: | ATP-binding cassette sub-family B member 6, mitochondrial, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Kim, S, Lee, S.S, Park, J.G, Kim, J.W, Kim, S, Kim, N.J, Hong, S, Kang, J.Y, Jin, M.S. | | Deposit date: | 2020-12-11 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Insights into Porphyrin Recognition by the Human ATP-Binding Cassette Transporter ABCB6.
Mol.Cells, 45, 2022
|
|
7DNY
 
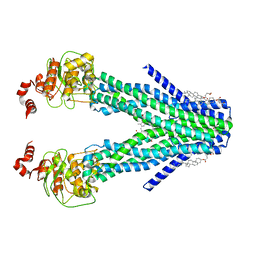 | | Cryo-EM structure of the human ABCB6 (coproporphyrin III-bound) | | Descriptor: | ATP-binding cassette sub-family B member 6, mitochondrial, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Kim, S, Lee, S.S, Park, J.G, Kim, J.W, Kim, S, Kim, N.J, Hong, S, Kang, J.Y, Jin, M.S. | | Deposit date: | 2020-12-11 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Insights into Porphyrin Recognition by the Human ATP-Binding Cassette Transporter ABCB6.
Mol.Cells, 45, 2022
|
|
5C6F
 
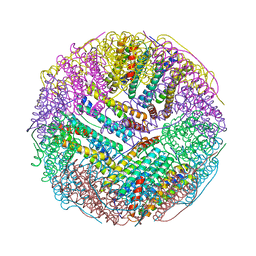 | | Crystal structures of ferritin mutants reveal side-on binding to diiron and end-on cleavage of oxygen | | Descriptor: | Bacterial non-heme ferritin, FE (III) ION, IMIDAZOLE | | Authors: | Kim, S, Kim, K.H, Seok, J.H, Park, Y.H, Jung, S.W, Chung, Y.B, Lee, D.B, Lee, J.H, Han, K.R, Cho, A.E, Lee, C, Chung, M.S. | | Deposit date: | 2015-06-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Novel Iron-Uptake Route and Reaction Intermediates in Ferritins from Gram-Negative Bacteria.
J. Mol. Biol., 428, 2016
|
|
4JDY
 
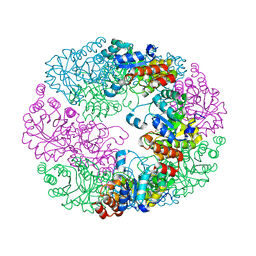 | | Crystal structure of Rv2606c | | Descriptor: | GLYCEROL, Pyridoxal biosynthesis lyase PdxS | | Authors: | Kim, S, Kim, K.-J. | | Deposit date: | 2013-02-25 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis Rv2606c: a pyridoxal biosynthesis lyase.
Biochem.Biophys.Res.Commun., 435, 2013
|
|
6KD7
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase | | Descriptor: | GLYCEROL, MAGNESIUM ION, PYROPHOSPHATE, ... | | Authors: | Kim, S, Kim, K.-J. | | Deposit date: | 2019-07-01 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of geranylgeranyl pyrophosphate synthase (crtE) from Nonlabens dokdonensis DSW-6.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
7VG9
 
 | |
6KIE
 
 | | Crystal structure of human leucyl-tRNA synthetase, Leu-AMS-bound form | | Descriptor: | 5'-O-(L-leucylsulfamoyl)adenosine, LEUCINE, Leucine--tRNA ligase, ... | | Authors: | Kim, S, Son, J, Kim, S, Hwang, K.Y. | | Deposit date: | 2019-07-18 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Leucine-sensing mechanism of leucyl-tRNA synthetase 1 for mTORC1 activation.
Cell Rep, 35, 2021
|
|
6KID
 
 | | Crystal structure of human leucyl-tRNA synthetase, ATP-bound form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, LEUCINE, Leucine--tRNA ligase, ... | | Authors: | Kim, S, Son, J, Kim, S, Hwang, K.Y. | | Deposit date: | 2019-07-18 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Leucine-sensing mechanism of leucyl-tRNA synthetase 1 for mTORC1 activation.
Cell Rep, 35, 2021
|
|
6KQY
 
 | | Crystal structure of human leucyl-tRNA synthetase, Leucine-bound form | | Descriptor: | LEUCINE, Leucine--tRNA ligase, cytoplasmic, ... | | Authors: | Kim, S, Son, J, Kim, S, Hwang, K.Y. | | Deposit date: | 2019-08-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Leucine-sensing mechanism of leucyl-tRNA synthetase 1 for mTORC1 activation.
Cell Rep, 35, 2021
|
|
6KR7
 
 | | Crystal structure of methylated human leucyl-tRNA synthetase, Leu-AMS-bound form | | Descriptor: | 5'-O-(L-leucylsulfamoyl)adenosine, LEUCINE, Leucine--tRNA ligase, ... | | Authors: | Kim, S, Son, J, Kim, S, Hwang, K.Y. | | Deposit date: | 2019-08-21 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Leucine-sensing mechanism of leucyl-tRNA synthetase 1 for mTORC1 activation.
Cell Rep, 35, 2021
|
|
4N45
 
 | | Crystal structure of reduced form of thiolase from Clostridium acetobutylicum | | Descriptor: | Acetyl-CoA acetyltransferase | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2013-10-08 | | Release date: | 2014-10-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insight into redox-switch regulatory mechanism of thiolase from the n-butanol synthesizing bacterium, Clostridium acetobutylicum
to be published
|
|
4N44
 
 | | Crystal structure of oxidized form of thiolase from Clostridium acetobutylicum | | Descriptor: | ACETATE ION, Acetyl-CoA acetyltransferase, GLYCEROL | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2013-10-08 | | Release date: | 2014-10-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural insight into redox-switch regulatory mechanism of thiolase from the n-butanol synthesizing bacterium, Clostridium acetobutylicum
to be published
|
|
4PRL
 
 | |
