8D3F
 
 | | Crystal structure of human STAT1 in complex with the repeat region from Toxoplasma protein TgIST | | Descriptor: | Signal transducer and activator of transcription 1-alpha/beta,Inhibitor of STAT1-dependent transcription TgIST | | Authors: | Huang, Z, Liu, H, Nix, J.C, Amarasinghe, G.K, Sibley, L.D. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | The intrinsically disordered protein TgIST from Toxoplasma gondii inhibits STAT1 signaling by blocking cofactor recruitment.
Nat Commun, 13, 2022
|
|
4QQ5
 
 | | Crystal Structure of FGF Receptor (FGFR) 4 Kinase Harboring the V550L Gate-Keeper Mutation in Complex With FIIN-2, an Irreversible Tyrosine Kinase Inhibitor Capable of Overcoming FGFR Kinase Gate-Keeper Mutations | | Descriptor: | Fibroblast growth factor receptor 4, N-(4-{[3-(3,5-dimethoxyphenyl)-7-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-oxo-3,4-dihydropyrimido[4,5-d]pyrimidin-1(2H)-yl]methyl}phenyl)propanamide | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-06-26 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | DFG-out Mode of Inhibition by an Irreversible Type-1 Inhibitor Capable of Overcoming Gate-Keeper Mutations in FGF Receptors.
Acs Chem.Biol., 10, 2015
|
|
4QQJ
 
 | |
4QQT
 
 | |
4QRC
 
 | | Crystal Structure of the Tyrosine Kinase Domain of FGF Receptor 4 in Complex with Ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Fibroblast growth factor receptor 4, SULFATE ION | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | DFG-out Mode of Inhibition by an Irreversible Type-1 Inhibitor Capable of Overcoming Gate-Keeper Mutations in FGF Receptors.
Acs Chem.Biol., 10, 2015
|
|
5EG3
 
 | | Crystal Structure of the Activated FGF Receptor 2 (FGFR2) Kinase Domain in complex with the cSH2 domain of Phospholipase C gamma (PLCgamma) | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1, Fibroblast growth factor receptor 2, MAGNESIUM ION, ... | | Authors: | Huang, Z, Li, X, Mohammadi, M. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Two FGF Receptor Kinase Molecules Act in Concert to Recruit and Transphosphorylate Phospholipase C gamma.
Mol.Cell, 61, 2016
|
|
4K33
 
 | | Crystal Structure of FGF Receptor 3 (FGFR3) Kinase Domain Harboring the K650E Mutation, a Gain-of-Function Mutation Responsible for Thanatophoric Dysplasia Type II and Spermatocytic Seminoma | | Descriptor: | Fibroblast growth factor receptor 3, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Huang, Z, Chen, H, Mohammadi, M. | | Deposit date: | 2013-04-10 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3405 Å) | | Cite: | Structural Mimicry of A-Loop Tyrosine Phosphorylation by a Pathogenic FGF Receptor 3 Mutation.
Structure, 21, 2013
|
|
8QAO
 
 | |
3DJA
 
 | | Crystal Structure of cpaf solved with MAD | | Descriptor: | Protein CT_858 | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-06-22 | | Release date: | 2009-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
3DPN
 
 | | Crystal Structure of cpaf s499a mutant | | Descriptor: | Protein CT_858 | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-07-09 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
3DOR
 
 | | Crystal Structure of mature CPAF | | Descriptor: | Protein CT_858, SULFATE ION | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-07-06 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
3DPM
 
 | | Structure of mature CPAF complexed with lactacystin | | Descriptor: | N-acetyl-S-({(2R,3S,4R)-3-hydroxy-2-[(1S)-1-hydroxy-2-methylpropyl]-4-methyl-5-oxopyrrolidin-2-yl}carbonyl)cysteine, Protein CT_858 | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-07-09 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
4R6V
 
 | | Crystal Structure of FGF Receptor (FGFR) 4 Kinase Harboring the V550L Gate-Keeper Mutation in Complex with FIIN-3, an Irreversible Tyrosine Kinase Inhibitor Capable of Overcoming FGFR kinase Gate-Keeper Mutations | | Descriptor: | Fibroblast growth factor receptor 4, N-[4-({[(2,6-dichloro-3,5-dimethoxyphenyl)carbamoyl](6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino}methyl)phenyl]propanamide, SULFATE ION | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3CXB
 
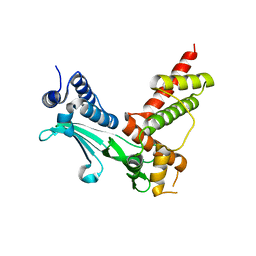 | | Crystal Structure of sifa and skip | | Descriptor: | Pleckstrin homology domain-containing family M member 2, Protein sifA | | Authors: | Huang, Z, Chai, J. | | Deposit date: | 2008-04-24 | | Release date: | 2008-12-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of Salmonella SifA indicate that its interactions with SKIP, SseJ, and RhoA family GTPases induce endosomal tubulation
Cell Host Microbe, 4, 2008
|
|
3GCG
 
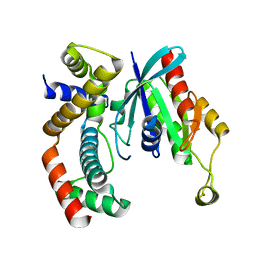 | | crystal structure of MAP and CDC42 complex | | Descriptor: | Cell division control protein 42 homolog, L0028 (Mitochondria associated protein) | | Authors: | Chai, J, Huang, Z, Feng, Y, Wu, X. | | Deposit date: | 2009-02-22 | | Release date: | 2009-07-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into host GTPase isoform selection by a family of bacterial GEF mimics
Nat.Struct.Mol.Biol., 16, 2009
|
|
1Z7I
 
 | |
4QQC
 
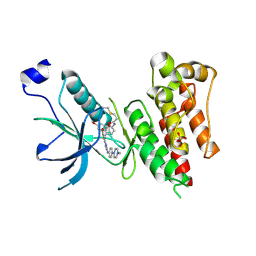 | | Crystal Structure of FGF Receptor (FGFR) 4 Kinase Domain in Complex with FIIN-2, an Irreversible Tyrosine Kinase Inhibitor Capable of Overcoming FGFR Kinase Gate-Keeper Mutations | | Descriptor: | Fibroblast growth factor receptor 4, N-(4-{[3-(3,5-dimethoxyphenyl)-7-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-oxo-3,4-dihydropyrimido[4,5-d]pyrimidin-1(2H)-yl]methyl}phenyl)propanamide, SULFATE ION | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-06-27 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7ZE9
 
 | | Structure of an AA16 LPMO-like protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (II) ION, ... | | Authors: | Huang, Z, Banerjee, S, Muderspach, S.J, Sun, P, van Berkel, W.J.H, Kabel, M.A, Lo Leggio, L. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.646 Å) | | Cite: | AA16 Oxidoreductases Boost Cellulose-Active AA9 Lytic Polysaccharide Monooxygenases from Myceliophthora thermophila.
Acs Catalysis, 13, 2023
|
|
4J98
 
 | |
5Y2F
 
 | | Human SIRT6 in complex with allosteric activator MDL-801 | | Descriptor: | 5-[[3,5-bis(chloranyl)phenyl]sulfonylamino]-2-[(5-bromanyl-4-fluoranyl-2-methyl-phenyl)sulfamoyl]benzoic acid, 9-mer peptide QTARKSTGG, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, J, Huang, Z, Song, K. | | Deposit date: | 2017-07-25 | | Release date: | 2018-11-07 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Identification of a cellularly active SIRT6 allosteric activator.
Nat. Chem. Biol., 14, 2018
|
|
3BM0
 
 | |
5W7O
 
 | |
5WJR
 
 | |
8YE6
 
 | |
5WTI
 
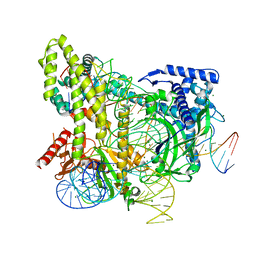 | | Crystal structure of the CRISPR-associated protein in complex with crRNA and DNA | | Descriptor: | CRISPR-associated protein, DNA (28-MER), DNA (5'-D(P*GP*TP*GP*TP*GP*GP*AP*TP*TP*CP*CP*G)-3'), ... | | Authors: | Wu, D, Guan, X, Zhu, Y, Huang, Z. | | Deposit date: | 2016-12-13 | | Release date: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.682 Å) | | Cite: | Structural basis of stringent PAM recognition by CRISPR-C2c1 in complex with sgRNA
Cell Res., 27, 2017
|
|
