5XRA
 
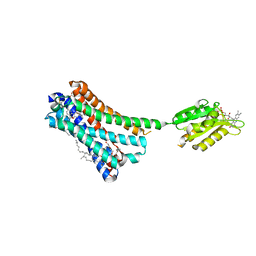 | | Crystal structure of the human CB1 in complex with agonist AM11542 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6aR,10aR)-3-(8-bromanyl-2-methyl-octan-2-yl)-6,6,9-trimethyl-6a,7,10,10a-tetrahydrobenzo[c]chromen-1-ol, CHOLESTEROL, ... | | Authors: | Hua, T, Vemuri, K, Nikas, P.S, Laprairie, R.B, Wu, Y, Qu, L, Pu, M, Korde, A, Shan, J, Ho, J.H, Han, G.W, Ding, K, Li, X, Liu, H, Hanson, M.A, Zhao, S, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of agonist-bound human cannabinoid receptor CB1
Nature, 547, 2017
|
|
5TGZ
 
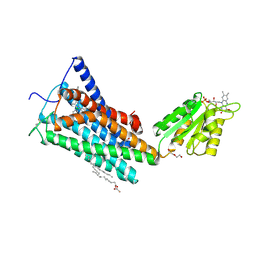 | | Crystal Structure of the Human Cannabinoid Receptor CB1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-[4-[2-(2,4-dichlorophenyl)-4-methyl-5-(piperidin-1-ylcarbamoyl)pyrazol-3-yl]phenyl]but-3-ynyl nitrate, Cannabinoid receptor 1,Flavodoxin,Cannabinoid receptor 1, ... | | Authors: | Hua, T, Vemuri, K, Pu, M, Qu, L, Han, G.W, Wu, Y, Zhao, S, Shui, W, Li, S, Korde, A, Laprairie, R.B, Stahl, E.L, Ho, J.H, Zvonok, N, Zhou, H, Kufareva, I, Wu, B, Zhao, Q, Hanson, M.A, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2016-09-28 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Human Cannabinoid Receptor CB1.
Cell, 167, 2016
|
|
6KPG
 
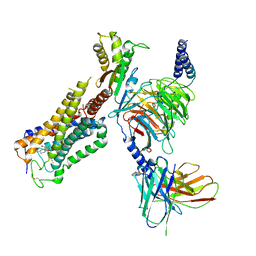 | | Cryo-EM structure of CB1-G protein complex | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hua, T, Li, X.T, Wu, L.J, Makriyannis, A, Wang, Y.X, Shen, L, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
5XR8
 
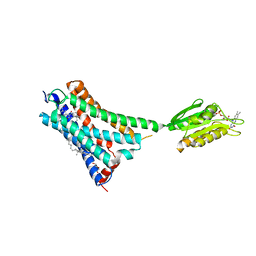 | | Crystal structure of the human CB1 in complex with agonist AM841 | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, CHOLESTEROL, Cannabinoid receptor 1,Flavodoxin,Cannabinoid receptor 1, ... | | Authors: | Hua, T, Vemuri, K, Nikas, P.S, Laprairie, R.B, Wu, Y, Qu, L, Pu, M, Korde, A, Shan, J, Ho, J.H, Han, G.W, Ding, K, Li, X, Liu, H, Hanson, M.A, Zhao, S, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structures of agonist-bound human cannabinoid receptor CB1.
Nature, 547, 2017
|
|
4FC7
 
 | | Studies on DCR shed new light on peroxisomal beta-oxidation: Crystal structure of the ternary complex of pDCR | | Descriptor: | COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Peroxisomal 2,4-dienoyl-CoA reductase | | Authors: | Hua, T, Wu, D, Wang, J, Shaw, N, Liu, Z.-J. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Studies of human 2,4-dienoyl CoA reductase shed new light on peroxisomal beta-oxidation of unsaturated fatty acids
J.Biol.Chem., 287, 2012
|
|
4FC6
 
 | | Studies on DCR shed new light on peroxisomal beta-oxidation: Crystal structure of the ternary complex of pDCR | | Descriptor: | HEXANOYL-COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Peroxisomal 2,4-dienoyl-CoA reductase | | Authors: | Hua, T, Wu, D, Wang, J, Shaw, N, Liu, Z.-J. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Studies of human 2,4-dienoyl CoA reductase shed new light on peroxisomal beta-oxidation of unsaturated fatty acids
J.Biol.Chem., 287, 2012
|
|
2MI8
 
 | |
6KPC
 
 | | Crystal structure of an agonist bound GPCR | | Descriptor: | 7-[(6aR,9R,10aR)-1-Hydroxy-9-(hydroxymethyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromen-3-yl]- 7-methyloctanenitrile, Cannabinoid receptor 2,Endolysin | | Authors: | Li, X.T, Hua, T, Wu, L.J, Makriyannis, A, Wu, M, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
6KPF
 
 | | Cryo-EM structure of a class A GPCR with G protein complex | | Descriptor: | 7-[(6aR,9R,10aR)-1-Hydroxy-9-(hydroxymethyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromen-3-yl]- 7-methyloctanenitrile, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, X.T, Hua, T, Wu, L.J, Makriyannis, A, Shen, L, Wang, Y.X, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
8X8O
 
 | | Cryo-EM structure of a bacteriophage tail- spike protein against Klebsiella pneumoniae K64,ORF41(K64-ORF41) in 5 mM EDTA | | Descriptor: | Probable tail spike protein | | Authors: | Xie, Y, Huang, T, Zhang, Z, Tao, X. | | Deposit date: | 2023-11-27 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural and functional basis of bacteriophage K64-ORF41 depolymerase for capsular polysaccharide degradation of Klebsiella pneumoniae K64.
Int.J.Biol.Macromol., 265, 2024
|
|
8X8M
 
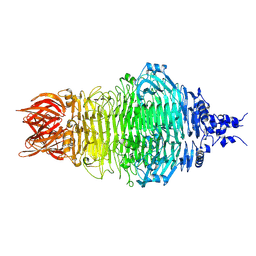 | | Cryo-EM structure of a bacteriophage tail- spike protein against Klebsiella pneumoniae K64,ORF41(K64-ORF41) | | Descriptor: | Probable tail spike protein | | Authors: | Xie, Y, Huang, T, Zhang, Z, Tao, X. | | Deposit date: | 2023-11-27 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Structural and functional basis of bacteriophage K64-ORF41 depolymerase for capsular polysaccharide degradation of Klebsiella pneumoniae K64.
Int.J.Biol.Macromol., 265, 2024
|
|
4NOJ
 
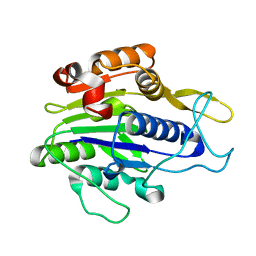 | | Crystal structure of the mature form of asparaginyl endopeptidase (AEP)/Legumain activated at pH 3.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
4NOL
 
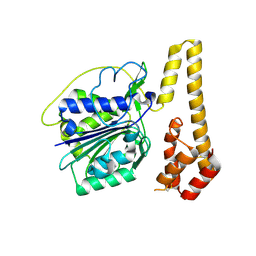 | | Crystal structure of proenzyme asparaginyl endopeptidase (AEP)/Legumain mutant D233A at pH 7.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
4NOK
 
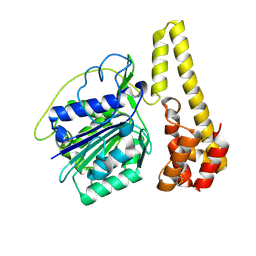 | | Crystal structure of proenzyme asparaginyl endopeptidase (AEP)/Legumain at pH 7.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2014-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
4NOM
 
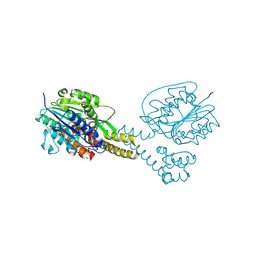 | | Crystal structure of asparaginyl endopeptidase (AEP)/Legumain activated at pH 4.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2014-03-19 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
8XQT
 
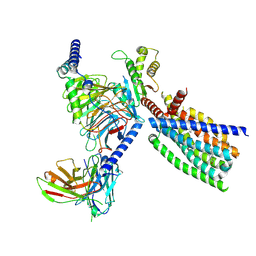 | | Structure of human class T GPCR TAS2R14-Gi complex. | | Descriptor: | CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hu, X.L, Pei, Y, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQR
 
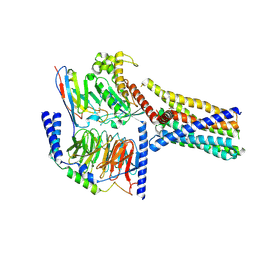 | | Structure 2 of human class T GPCR TAS2R14-miniGs/gust complex with Flufenamic acid. | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQO
 
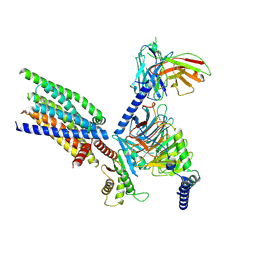 | | Structure of human class T GPCR TAS2R14-Gi complex with Aristolochic acid A. | | Descriptor: | 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQS
 
 | | Structure of human class T GPCR TAS2R14-DNGi complex with Flufenamic acid. | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XVJ
 
 | | Cryo-EM structure of ETAR bound with Macitentan | | Descriptor: | 6-[2-(5-bromanylpyrimidin-2-yl)oxyethoxy]-5-(4-bromophenyl)-~{N}-(propylsulfamoyl)pyrimidin-4-amine, Endoglucanase H,Endothelin-1 receptor,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVK
 
 | | Cryo-EM structure of ETAR bound with Ambrisentan | | Descriptor: | (2~{S})-2-(4,6-dimethylpyrimidin-2-yl)oxy-3-methoxy-3,3-diphenyl-propanoic acid, Endoglucanase H,Endothelin-1 receptor,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVL
 
 | | Cryo-EM structure of ETAR bound with Zibotentan | | Descriptor: | Endoglucanase H,Endothelin-1 receptor,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, anti-BRIL Fab Light chain, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVI
 
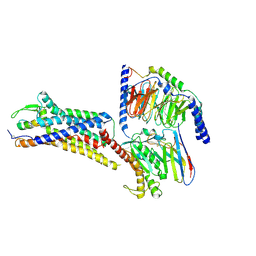 | | Cryo-EM structure of ETAR bound with Endothelin1 | | Descriptor: | Endoglucanase H,Endothelin-1 receptor, Endothelin-1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVH
 
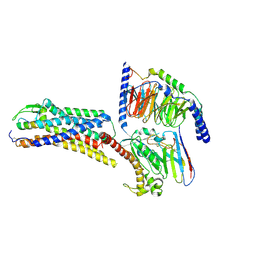 | | Cryo-EM structure of ETBR bound with Endothelin1 | | Descriptor: | Endothelin-1, Exo-alpha-sialidase,Endothelin receptor type B, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVE
 
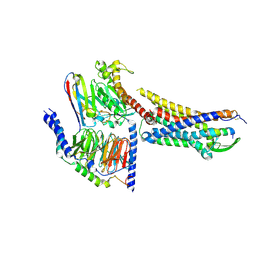 | | Cryo-EM structure of ETBR bound with BQ3020 | | Descriptor: | BQ3020, Exo-alpha-sialidase,Endothelin receptor type B, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
