7DCU
 
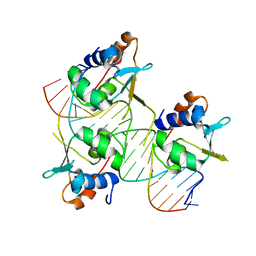 | | Crystal structure of HSF2 DNA-binding domain in complex with 3-site HSE DNA (21 bp) | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*CP*GP*AP*AP*TP*AP*TP*TP*CP*TP*AP*GP*AP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*TP*CP*TP*AP*GP*AP*AP*TP*AP*TP*TP*CP*GP*CP*GP*G)-3'), Heat shock factor protein 2, ... | | Authors: | Feng, N, Liu, W. | | Deposit date: | 2020-10-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of heat shock factor trimers bound to DNA.
Iscience, 24, 2021
|
|
7DCT
 
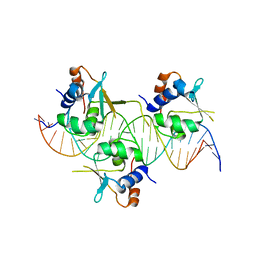 | | Crystal structure of HSF1 DNA-binding domain in complex with 3-site HSE DNA (24 bp) | | Descriptor: | DNA (5'-D(*AP*CP*TP*CP*GP*CP*GP*AP*AP*TP*AP*TP*TP*CP*TP*AP*GP*AP*AP*CP*GP*CP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*GP*TP*TP*CP*TP*AP*GP*AP*AP*TP*AP*TP*TP*CP*GP*CP*GP*AP*G)-3'), Heat shock factor protein 1, ... | | Authors: | Feng, N, Liu, W. | | Deposit date: | 2020-10-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structures of heat shock factor trimers bound to DNA.
Iscience, 24, 2021
|
|
7DCJ
 
 | |
7DCS
 
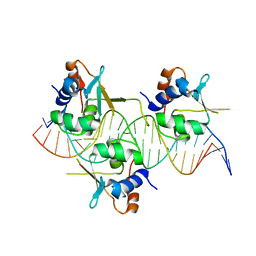 | | Crystal structure of HSF1 DNA-binding domain in complex with 3-site HSE DNA (23 bp) | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*GP*CP*GP*AP*AP*TP*AP*TP*TP*CP*TP*AP*GP*AP*AP*CP*GP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*CP*GP*TP*TP*CP*TP*AP*GP*AP*AP*TP*AP*TP*TP*CP*GP*CP*GP*GP*A)-3'), Heat shock factor protein 1, ... | | Authors: | Feng, N, Liu, W. | | Deposit date: | 2020-10-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of heat shock factor trimers bound to DNA.
Iscience, 24, 2021
|
|
5YZ2
 
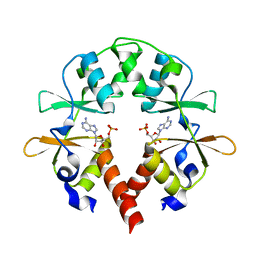 | | the cystathionine-beta-synthase (CBS) domain of magnesium and cobalt efflux protein CorC in complex with both C2'- and C3'-endo AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Magnesium and cobalt efflux protein CorC | | Authors: | Feng, N, Qi, C, Li, D.F, Wang, D.C. | | Deposit date: | 2017-12-12 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The C2'- and C3'-endo equilibrium for AMP molecules bound in the cystathionine-beta-synthase domain.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
7XPY
 
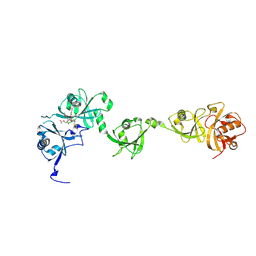 | | Crystal structure of USP7 in complex with its inhibitor | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7, [(3S,3aR,4R,6Z,9S,10E,11aR)-9-acetyloxy-6-(acetyloxymethyl)-3,10-dimethyl-2-oxidanylidene-3a,4,5,8,9,11a-hexahydro-3H-cyclodeca[b]furan-4-yl] (E)-2-methyl-4-oxidanyl-but-2-enoate | | Authors: | Feng, N, Zeng, K.W. | | Deposit date: | 2022-05-06 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Neuroinflammation inhibition by small-molecule targeting USP7 noncatalytic domain for neurodegenerative disease therapy.
Sci Adv, 8, 2022
|
|
5HAA
 
 | |
7VIJ
 
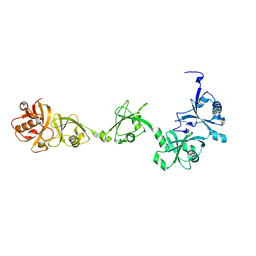 | | Crystal structure of USP7-HUBL domain | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Feng, N, Zeng, K.W. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Neuroinflammation inhibition by small-molecule targeting USP7 noncatalytic domain for neurodegenerative disease therapy.
Sci Adv, 8, 2022
|
|
4YIF
 
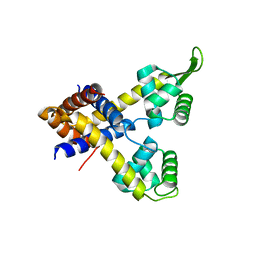 | | Crystal structure of Rv0880 | | Descriptor: | MarR family protein Rv0880 | | Authors: | Gao, Y.R, Feng, N, Li, D.F, Bi, L.J. | | Deposit date: | 2015-03-02 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure of the MarR family protein Rv0880 from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5WS2
 
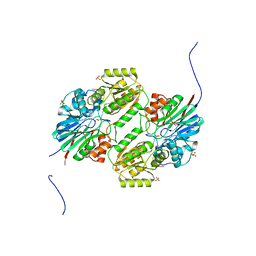 | | Crystal structure of mpy-RNase J (mutant S247A), an archaeal RNase J from Methanolobus psychrophilus R15, complex with RNA | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*A)-3'), Ribonuclease J, SULFATE ION, ... | | Authors: | Li, D.F, Feng, N. | | Deposit date: | 2016-12-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | New molecular insights into an archaeal RNase J reveal a conserved processive exoribonucleolysis mechanism of the RNase J family
Mol. Microbiol., 106, 2017
|
|
5HAB
 
 | |
6PCU
 
 | |
7R75
 
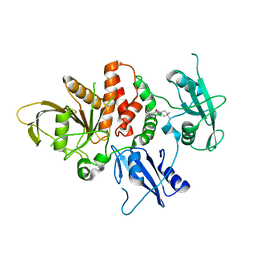 | | Structure of human SHP2 in complex with compound 16 | | Descriptor: | 6-(4-amino-4-methylpiperidin-1-yl)-3-(3-chlorophenyl)-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Leonard, P.G, Cross, J. | | Deposit date: | 2021-06-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Discovery of 6-[(3 S ,4 S )-4-Amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-(2,3-dichlorophenyl)-2-methyl-3,4-dihydropyrimidin-4-one (IACS-15414), a Potent and Orally Bioavailable SHP2 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7R7D
 
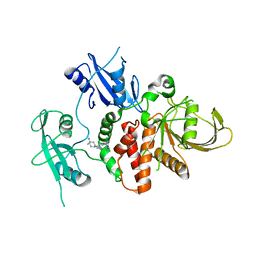 | | Structure of human SHP2 in complex with compound 22 | | Descriptor: | 4-[6-(4-amino-4-methylpiperidin-1-yl)-1H-pyrazolo[3,4-b]pyrazin-3-yl]-3-chloro-N-methylpyridin-2-amine, TETRAETHYLENE GLYCOL, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Leonard, P.G, Cross, J. | | Deposit date: | 2021-06-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 6-[(3 S ,4 S )-4-Amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-(2,3-dichlorophenyl)-2-methyl-3,4-dihydropyrimidin-4-one (IACS-15414), a Potent and Orally Bioavailable SHP2 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7R7L
 
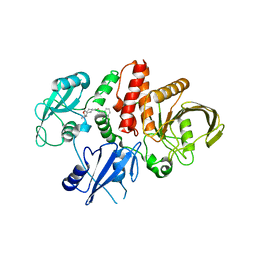 | | Structure of human SHP2 in complex with compound 30 | | Descriptor: | 6-[(3S,4S)-4-amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-(2,3-dichlorophenyl)-2-methylpyrimidin-4(3H)-one, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Leonard, P.G, Cross, J. | | Deposit date: | 2021-06-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of 6-[(3 S ,4 S )-4-Amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-(2,3-dichlorophenyl)-2-methyl-3,4-dihydropyrimidin-4-one (IACS-15414), a Potent and Orally Bioavailable SHP2 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7R7I
 
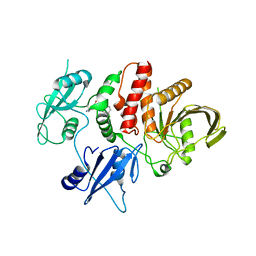 | | Structure of human SHP2 in complex with compound 27 | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11, [3-(4-amino-4-methylpiperidin-1-yl)-6-(2,3-dichlorophenyl)-5-methylpyrazin-2-yl]methanol | | Authors: | Leonard, P.G, Cross, J. | | Deposit date: | 2021-06-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of 6-[(3 S ,4 S )-4-Amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-(2,3-dichlorophenyl)-2-methyl-3,4-dihydropyrimidin-4-one (IACS-15414), a Potent and Orally Bioavailable SHP2 Inhibitor.
J.Med.Chem., 64, 2021
|
|
6WU8
 
 | | Structure of human SHP2 in complex with inhibitor IACS-13909 | | Descriptor: | 1-[3-(2,3-dichlorophenyl)-1H-pyrazolo[3,4-b]pyrazin-6-yl]-4-methylpiperidin-4-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Leonard, P.G, Joseph, S, Rodenberger, A. | | Deposit date: | 2020-05-04 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric SHP2 Inhibitor, IACS-13909, Overcomes EGFR-Dependent and EGFR-Independent Resistance Mechanisms toward Osimertinib.
Cancer Res., 80, 2020
|
|
5UQ9
 
 | | Crystal structure of 6-phosphogluconate dehydrogenase with ((4R,5R)-5-(hydroxycarbamoyl)-2,2-dimethyl-1,3-dioxolan-4-yl)methyl dihydrogen phosphate | | Descriptor: | 6-phosphogluconate dehydrogenase, decarboxylating, [(4R,5R)-5-(hydroxycarbamoyl)-2,2-dimethyl-1,3-dioxolan-4-yl]methyl dihydrogen phosphate | | Authors: | Leonard, P.G. | | Deposit date: | 2017-02-07 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional Genomics Reveals Synthetic Lethality between Phosphogluconate Dehydrogenase and Oxidative Phosphorylation.
Cell Rep, 26, 2019
|
|
7WR5
 
 | | Crystal structure of OspC3-calmodulin-caspase-4 complex binding with 2'-aF-NAD+ | | Descriptor: | Calmodulin-1, Caspase-4, OspC3, ... | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR4
 
 | | Crystal structure of OspC3-calmodulin-caspase-4 complex | | Descriptor: | Calmodulin-1, Caspase-4, OspC3 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR0
 
 | | P32 of caspase-4 C258A mutant | | Descriptor: | Caspase-4 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR2
 
 | |
7WR6
 
 | | Crystal structure of ADP-riboxanated caspase-4 in complex with Af1521 | | Descriptor: | ADP-ribose glycohydrolase AF_1521, Caspase-4, [[(3~{a}~{S},5~{R},6~{R},6~{a}~{R})-2-azanylidene-3-[(4~{R})-4-azanyl-5-oxidanylidene-pentyl]-6-oxidanyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]oxazol-5-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR1
 
 | | P32 of caspase-4 C258A mutant in complex with OspC3 C-terminal ankyrin-repeat domain | | Descriptor: | Caspase-4, OspC3 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR3
 
 | | Crystal structure of MBP-fused OspC3 in complex with calmodulin | | Descriptor: | Calmodulin-1, MBP-fused OspC3, NICOTINAMIDE, ... | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
