4RYL
 
 | | Human Protein Arginine Methyltransferase 3 in complex with 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea | | Descriptor: | 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea, PRMT3 protein, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Dobrovetsky, E, Kaniskan, H.U, Szewczyk, M, Yu, Z, Eram, M.S, Yang, X, Schmidt, K, Luo, X, Dai, M, He, F, Zang, I, Lin, Y, Kennedy, S, Li, F, Tempel, W, Smil, D, Min, S.J, Landon, M, Lin-Jones, J, Huang, X.P, Roth, B.L, Schapira, M, Atadja, P, Barsyte-Lovejoy, D, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Zhao, K, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent, Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 3 (PRMT3).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4QQN
 
 | | Protein arginine methyltransferase 3 in complex with compound MTV044246 | | Descriptor: | 1-{2-[1-(aminomethyl)cyclohexyl]ethyl}-3-isoquinolin-6-ylurea, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dong, A, Dobrovetsky, E, Tempel, W, He, H, Zhao, K, Smil, D, Landon, M, Luo, X, Chen, Z, Dai, M, Yu, Z, Lin, Y, Zhang, H, Zhao, K, Schapira, M, Brown, P.J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of Potent and Selective Allosteric Inhibitors of Protein Arginine Methyltransferase 3 (PRMT3).
J. Med. Chem., 61, 2018
|
|
6NB5
 
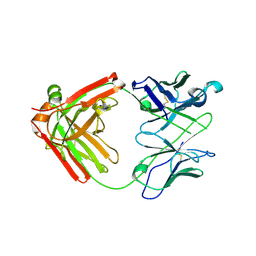 | | Crystal structure of anti- MERS-CoV human neutralizing LCA60 antibody Fab fragment | | Descriptor: | LCA60 antigen-binding (Fab) fragment, heavy chain, light chain | | Authors: | Walls, A.J, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, J, Quispe, J, Cameroni, E, Gopal, R, Dai, M, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NB8
 
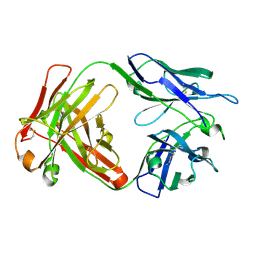 | | Crystal structure of anti- SARS-CoV human neutralizing S230 antibody Fab fragment | | Descriptor: | S230 antigen-binding (Fab) fragment, heavy chain, light chain | | Authors: | Walls, A.J, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, J, Quispe, J, Cameroni, E, Gopal, R, Dai, M, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
7E6T
 
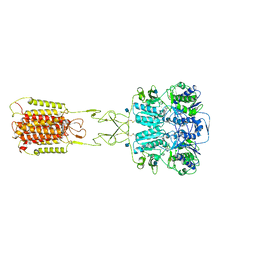 | | Structural insights into the activation of human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYCLOMETHYLTRYPTOPHAN, ... | | Authors: | Geng, Y, Chen, X.C, Wang, L, Cui, Q.Q, Ding, Z.Y, Han, L, Kou, Y.J, Zhang, W.Q, Wang, H.N, Jia, X.M, Dai, M, Shi, Z.Z, Li, Y.Y, Li, X.Y. | | Deposit date: | 2021-02-24 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the activation of human calcium-sensing receptor.
Elife, 10, 2021
|
|
7E6U
 
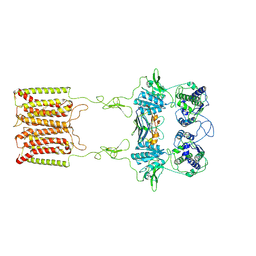 | | the complex of inactive CaSR and NB2D11 | | Descriptor: | Extracellular calcium-sensing receptor, NB-2D11 | | Authors: | Geng, Y, Chen, X.C, Wang, L, Cui, Q.Q, Ding, Z.Y, Han, L, Kou, Y.J, Zhang, W.Q, Wang, H.N, Jia, X.M, Dai, M, Shi, Z.Z, Li, Y.Y, Li, X.Y. | | Deposit date: | 2021-02-24 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural insights into the activation of human calcium-sensing receptor.
Elife, 10, 2021
|
|
1TOR
 
 | | MOLECULAR DYNAMICS SIMULATION FROM 2D-NMR DATA OF THE FREE ACHR MIR DECAPEPTIDE AND THE ANTIBODY-BOUND [A76]MIR ANALOGUE | | Descriptor: | ACETYLCHOLINE RECEPTOR, MAIN IMMUNOGENIC REGION | | Authors: | Orlewski, P, Tsikaris, V, Sakarellos, C, Sakarellos-Daistiotis, M, Vatzaki, E, Tzartos, S.J, Marraud, M, Cung, M.T. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Compared structures of the free nicotinic acetylcholine receptor main immunogenic region (MIR) decapeptide and the antibody-bound [A76]MIR analogue: a molecular dynamics simulation from two-dimensional NMR data.
Biopolymers, 40, 1996
|
|
1TOS
 
 | | TORPEDO CALIFORNICA ACHR RECEPTOR [ALA76] ANALOGUE COMPLEXED WITH THE ANTI-ACETYLCHOLINE MAB6 MONOCLONAL ANTIBODY | | Descriptor: | ACETYLCHOLINE RECEPTOR [ALA76] MIR ANALOGUE | | Authors: | Orlewski, P, Tsikaris, V, Sakarellos, C, Sakarellos-Daistiotis, M, Vatzaki, E, Tzartos, S.J, Marraud, M, Cung, M.T. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Compared structures of the free nicotinic acetylcholine receptor main immunogenic region (MIR) decapeptide and the antibody-bound [A76]MIR analogue: a molecular dynamics simulation from two-dimensional NMR data.
Biopolymers, 40, 1996
|
|
1F3R
 
 | | COMPLEX BETWEEN FV ANTIBODY FRAGMENT AND AN ANALOGUE OF THE MAIN IMMUNOGENIC REGION OF THE ACETYLCHOLINE RECEPTOR | | Descriptor: | ACETYLCHOLINE RECEPTOR ALPHA, FV ANTIBODY FRAGMENT | | Authors: | Kleinjung, J, Petit, M.-C, Orlewski, P, Mamalaki, A, Tzartos, S.-J, Tsikaris, V, Sakarellos-Daitsiotis, M, Sakarellos, C, Marraud, M, Cung, M.-T. | | Deposit date: | 2000-06-06 | | Release date: | 2000-06-15 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | The third-dimensional structure of the complex between an Fv antibody fragment and an analogue of the main immunogenic region of the acetylcholine receptor: a combined two-dimensional NMR, homology, and molecular modeling approach.
Biopolymers, 53, 2000
|
|
6GWG
 
 | | Alpha-galactosidase from Thermotoga maritima in complex with cyclohexene-based carbasugar mimic of galactose covalently linked to the nucleophile | | Descriptor: | (1~{S},2~{S},3~{S})-3-fluoranyl-6-(hydroxymethyl)cyclohex-5-ene-1,2,4-triol, Alpha-galactosidase, GLYCEROL, ... | | Authors: | Gloster, T.M, Oehler, V. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Revealing the mechanism for covalent inhibition of glycoside hydrolases by carbasugars at an atomic level.
Nat Commun, 9, 2018
|
|
6GVD
 
 | | Alpha-galactosidase from Thermotoga maritima in complex with cyclohexene-based carbasugar mimic of galactose | | Descriptor: | (1~{S},2~{S},3~{S},4~{S})-5-(hydroxymethyl)cyclohex-5-ene-1,2,3,4-tetrol, Alpha-galactosidase, MAGNESIUM ION, ... | | Authors: | Gloster, T.M, Pengelly, R.J. | | Deposit date: | 2018-06-20 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Revealing the mechanism for covalent inhibition of glycoside hydrolases by carbasugars at an atomic level.
Nat Commun, 9, 2018
|
|
6GX8
 
 | | Alpha-galactosidase from Thermotoga maritima in complex with hydrolysed cyclohexene-based carbasugar mimic of galactose | | Descriptor: | (1~{S},2~{S},3~{S},4~{S})-3-fluoranyl-6-(hydroxymethyl)cyclohex-5-ene-1,2,4-triol, Alpha-galactosidase, GLYCEROL, ... | | Authors: | Gloster, T.M, Oehler, V. | | Deposit date: | 2018-06-26 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Revealing the mechanism for covalent inhibition of glycoside hydrolases by carbasugars at an atomic level.
Nat Commun, 9, 2018
|
|
6GTA
 
 | | Alpha-galactosidase mutant D378A from Thermotoga maritima in complex with intact cyclohexene-based carbasugar mimic of galactose with 3,5 difluorophenyl leaving group | | Descriptor: | (1~{R},2~{S},3~{S},6~{S})-6-[3,5-bis(fluoranyl)phenoxy]-4-(hydroxymethyl)cyclohex-4-ene-1,2,3-triol, Alpha-galactosidase, MAGNESIUM ION, ... | | Authors: | Gloster, T.M, Pengelly, R.J. | | Deposit date: | 2018-06-17 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Revealing the mechanism for covalent inhibition of glycoside hydrolases by carbasugars at an atomic level.
Nat Commun, 9, 2018
|
|
6GWF
 
 | | Alpha-galactosidase mutant D387A from Thermotoga maritima in complex with intact cyclohexene-based carbasugar mimic of galactose with 2,4-dinitro leaving group | | Descriptor: | (1~{S},2~{S},5~{S},6~{R})-5-(2,4-dinitrophenoxy)-6-fluoranyl-3-(hydroxymethyl)cyclohex-3-ene-1,2-diol, Alpha-galactosidase, MAGNESIUM ION, ... | | Authors: | Gloster, T.M, Oehler, V. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Revealing the mechanism for covalent inhibition of glycoside hydrolases by carbasugars at an atomic level.
Nat Commun, 9, 2018
|
|
6NB4
 
 | | MERS-CoV S complex with human neutralizing LCA60 antibody Fab fragment (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LCA60 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NB7
 
 | | SARS-CoV complex with human neutralizing S230 antibody Fab fragment (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S230 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NB6
 
 | | SARS-CoV complex with human neutralizing S230 antibody Fab fragment (state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S230 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NB3
 
 | | MERS-CoV complex with human neutralizing LCA60 antibody Fab fragment (state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LCA60 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
7V40
 
 | | Crystal structure of Class I P450 monooxygenase (P450tol) from Rhodococcus coprophilus TC-2 | | Descriptor: | PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, p450tol monooxygenase | | Authors: | Zhang, L.L, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Molecular Basis for a Toluene Monooxygenase to Govern Substrate Selectivity
Acs Catalysis, 12, 2022
|
|
7V41
 
 | | Crystal structure of Class I P450 monooxygenase (P450tol) from Rhodococcus coprophilus TC-2 in complex with toluene. | | Descriptor: | PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, TOLUENE, ... | | Authors: | Zhang, L.L, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular Basis for a Toluene Monooxygenase to Govern Substrate Selectivity
Acs Catalysis, 12, 2022
|
|
7V44
 
 | | Crystal structure of Class I P450 monooxygenase (P450tol) from Rhodococcus coprophilus TC-2 in complex with meta-chlorotoluene. | | Descriptor: | 1-chloranyl-2-methyl-benzene, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Zhang, L.L, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Molecular Basis for a Toluene Monooxygenase to Govern Substrate Selectivity
Acs Catalysis, 12, 2022
|
|
7V43
 
 | | Crystal structure of Class I P450 monooxygenase (P450tol) from Rhodococcus coprophilus TC-2 in complex with para-chlorotoluene. | | Descriptor: | 1-chloranyl-4-methyl-benzene, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Zhang, L.L, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Basis for a Toluene Monooxygenase to Govern Substrate Selectivity
Acs Catalysis, 12, 2022
|
|
7V42
 
 | | Crystal structure of Class I P450 monooxygenase (P450tol) from Rhodococcus coprophilus TC-2 in complex with benzyl-alcohol. | | Descriptor: | PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, p450tol monooxygenase, ... | | Authors: | Zhang, L.L, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular Basis for a Toluene Monooxygenase to Govern Substrate Selectivity
Acs Catalysis, 12, 2022
|
|
7V45
 
 | | Crystal structure of Class I P450 monooxygenase (P450tol) from Rhodococcus coprophilus TC-2 in complex with para-bromotoluene. | | Descriptor: | 1-bromanyl-4-methyl-benzene, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, L.L, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular Basis for a Toluene Monooxygenase to Govern Substrate Selectivity
Acs Catalysis, 12, 2022
|
|
7V46
 
 | | Crystal structure of Class I P450 monooxygenase (P450tol) from Rhodococcus coprophilus TC-2 in complex with ortho-chlorotoluene. | | Descriptor: | 1-chloranyl-3-methyl-benzene, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, L.L, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for a Toluene Monooxygenase to Govern Substrate Selectivity
Acs Catalysis, 12, 2022
|
|
