6TTR
 
 | | Crystal Structure of the coiled coil and GGDEF domain of DgcB from Caulobacter crescentus in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), GGDEF diguanylate cyclase DgcB, PHOSPHATE ION | | Authors: | Holzschuh, F, Schirmer, T, Teixeira, R.D. | | Deposit date: | 2019-12-30 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the coiled coil and GGDEF domain of DgcB from Caulobacter crescentus in complex with c-di-GMP
To Be Published
|
|
6TTS
 
 | | Crystal structure of the GGDEF domain of DgcB from Caulobacter crescentus in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), GGDEF diguanylate cyclase DgcB, SULFATE ION | | Authors: | Holzschuh, F, Schirmer, T, Teixeira, R. | | Deposit date: | 2019-12-30 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the GGDEF domain of DgcB from Caulobacter crescentus in complex with c-di-GMP
To Be Published
|
|
1ZVS
 
 | | Crystal structure of the first class MHC mamu and Tat-Tl8 complex | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Tat-Tl8 | | Authors: | Lou, Z, Chu, F, Gao, G.F, Rao, Z. | | Deposit date: | 2005-06-02 | | Release date: | 2006-06-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | First glimpse of the peptide presentation by rhesus macaque MHC class I: crystal structures of Mamu-A*01 complexed with two immunogenic SIV epitopes and insights into CTL escape.
J Immunol., 178, 2007
|
|
3I6K
 
 | | Newly identified epitope from SARS-CoV membrane protein complexed with HLA-A*0201 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Liu, J, Sun, Y, Qi, J, Chu, F, Wu, H, Gao, F, Li, T, Yan, J, Gao, G.F. | | Deposit date: | 2009-07-07 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes
J Infect Dis, 202, 2010
|
|
7DS7
 
 | | The Crystal Structure of Leaf-branch compost cutinase from Biortus. | | Descriptor: | CITRIC ACID, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wang, F, Lv, Z, Cheng, W, Lin, D, Chu, F, Xu, X, Tan, J. | | Deposit date: | 2020-12-30 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of Leaf-branch compost cutinase from Biortus.
To Be Published
|
|
2K13
 
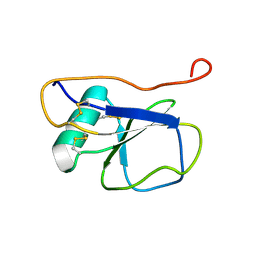 | | Solution NMR Structure of the Leech Protein Saratin, a Novel Inhibitor of Haemostasis | | Descriptor: | Saratin | | Authors: | Gronwald, W, Bomke, J, Maurer, T, Wisotzki, B, Huber, F, Schumann, F, Kremer, W, Frech, M, Kalbitzer, H.R. | | Deposit date: | 2008-02-20 | | Release date: | 2008-10-21 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of the leech protein saratin and characterization of its binding to collagen
J.Mol.Biol., 381, 2008
|
|
6Y20
 
 | |
3GH3
 
 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, Ecto-NAD+ glycohydrolase (CD38 molecule), ... | | Authors: | Egea, P.F, Muller-Steffner, H, Stroud, R.M, Kellenberger, E, Oppenheimer, N, Schuber, F. | | Deposit date: | 2009-03-02 | | Release date: | 2010-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
3GC6
 
 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ecto-NAD+ glycohydrolase (CD38 molecule), SULFATE ION | | Authors: | Egea, P.F, Muller-Steffner, H, Stroud, R.M, Oppenheimer, N, Kellenberger, E, Schuber, F. | | Deposit date: | 2009-02-21 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
3GHH
 
 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ecto-NAD+ glycohydrolase (CD38 molecule), SULFATE ION, ... | | Authors: | Egea, P.F, Muller-Steffner, H, Stroud, R.M, Oppenheimer, N.J, Kellenberger, E, Schuber, F. | | Deposit date: | 2009-03-03 | | Release date: | 2010-03-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
3KOU
 
 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD38 molecule, ... | | Authors: | Egea, P.F, Muller-Steffner, H, Stroud, R.M, Oppenheimer, N.J, Kellenberger, E, Schuber, F. | | Deposit date: | 2009-11-13 | | Release date: | 2010-10-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
3P5S
 
 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD38 molecule, SULFATE ION, ... | | Authors: | Egea, P.F, Muller-Stauffler, H, Kohn, I, Cakou-Kefir, C, Stroud, R.M, Kellenberburger, E, Schuber, F. | | Deposit date: | 2010-10-10 | | Release date: | 2011-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
4CH9
 
 | | Crystal structure of the human KLHL3 Kelch domain in complex with a WNK4 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, KELCH-LIKE PROTEIN 3, ... | | Authors: | Sorrell, F.J, Schumacher, F.R, Kurz, T, Alessi, D.R, Newman, J, Goubin, S, Chalk, R, Kopec, J, Tallant, C, Williams, E, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2013-11-29 | | Release date: | 2014-01-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural and Biochemical Characterisation of the Klhl3-Wnk Kinase Interaction Important in Blood Pressure Regulation.
Biochem.J., 460, 2014
|
|
4CHB
 
 | | Crystal structure of the human KLHL2 Kelch domain in complex with a WNK4 peptide | | Descriptor: | 1,2-ETHANEDIOL, DODECAETHYLENE GLYCOL, KELCH-LIKE PROTEIN 2, ... | | Authors: | Sorrell, F.J, Schumacher, F.R, Kurz, T, Alessi, D.R, Newman, J, Cooper, C.D.O, Canning, P, Kopec, J, Williams, E, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2013-11-29 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and Biochemical Characterisation of the Klhl3-Wnk Kinase Interaction Important in Blood Pressure Regulation.
Biochem.J., 460, 2014
|
|
6SEO
 
 | | TEAD4 bound to a FAM181B peptide | | Descriptor: | Protein FAM181B, Transcriptional enhancer factor TEF-3 | | Authors: | Scheufler, C, Villard, F. | | Deposit date: | 2019-07-30 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification of FAM181A and FAM181B as new interactors with the TEAD transcription factors.
Protein Sci., 29, 2020
|
|
6SEN
 
 | | TEAD4 bound to a FAM181A peptide | | Descriptor: | Protein FAM181A, SULFATE ION, Transcriptional enhancer factor TEF-3 | | Authors: | Scheufler, C, Villard, F. | | Deposit date: | 2019-07-30 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of FAM181A and FAM181B as new interactors with the TEAD transcription factors.
Protein Sci., 29, 2020
|
|
6HIL
 
 | |
6HIK
 
 | |
3FN3
 
 | | Dimeric Structure of PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Chen, Y, Gao, F, Liu, P, Chu, F, Qi, J, Gao, G.F. | | Deposit date: | 2008-12-23 | | Release date: | 2009-12-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A dimeric structure of PD-L1: functional units or evolutionary relics?
Protein Cell, 1, 2010
|
|
4K7P
 
 | |
6X88
 
 | | PDE6 chicken GAF domain | | Descriptor: | Cone cGMP-specific 3',5'-cyclic phosphodiesterase subunit alpha' | | Authors: | Ke, H. | | Deposit date: | 2020-06-01 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1997447 Å) | | Cite: | Structural Analysis of the Regulatory GAF Domains of cGMP Phosphodiesterase Elucidates the Allosteric Communication Pathway.
J.Mol.Biol., 432, 2020
|
|
3I6G
 
 | | Newly identified epitope Mn2 from SARS-CoV M protein complexed withHLA-A*0201 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Liu, J. | | Deposit date: | 2009-07-07 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes.
J.INFECT.DIS., 202, 2010
|
|
4GL2
 
 | | Structural Basis for dsRNA duplex backbone recognition by MDA5 | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RNA (5'-R(*AP*UP*CP*CP*GP*CP*GP*GP*CP*CP*CP*U)-3'), ... | | Authors: | Wu, B, Hur, S. | | Deposit date: | 2012-08-13 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.557 Å) | | Cite: | Structural Basis for dsRNA Recognition, Filament Formation, and Antiviral Signal Activation by MDA5.
Cell(Cambridge,Mass.), 152, 2013
|
|
4WV7
 
 | | HEAT SHOCK PROTEIN 70 SUBSTRATE BINDING DOMAIN WITH COVALENTLY LINKED NOVOLACTONE | | Descriptor: | (5beta,6alpha,8alpha,14alpha)-13-ethenyl-5,6-dihydroxy-14-methylpodocarp-12-en-15-oic acid, Heat shock 70 kDa protein 1A/1B | | Authors: | Kirby, C.A, Baird, J, Stams, T. | | Deposit date: | 2014-11-04 | | Release date: | 2015-01-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The novolactone natural product disrupts the allosteric regulation of hsp70.
Chem.Biol., 22, 2015
|
|
4WV5
 
 | |
