7ES4
 
 | | the crystral structure of DndH-C-domain | | Descriptor: | DNA phosphorothioation-dependent restriction protein DptH | | Authors: | Wu, D. | | Deposit date: | 2021-05-08 | | Release date: | 2022-05-11 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The functional coupling between restriction and DNA phosphorothioate modification systems underlying the DndFGH restriction complex
Nat Catal, 2022
|
|
7X6Z
 
 | |
7X70
 
 | |
7X6Y
 
 | |
7EXX
 
 | | The structure of DndG | | Descriptor: | DNA phosphorothioation-dependent restriction protein DptG, SODIUM ION | | Authors: | Wu, D, Wang, L. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-01 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The functional coupling between restriction and DNA phosphorothioate modification systems underlying the DndFGH restriction complex
Nat Catal, 2022
|
|
7WJQ
 
 | | Crystal structure of GSDMB in complex with Ipah7.8 | | Descriptor: | Isoform 2 of Gasdermin-B, Probable E3 ubiquitin-protein ligase ipaH7.8 | | Authors: | Li, X, Zhang, H, Yin, H. | | Deposit date: | 2022-01-07 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the GSDMB-mediated cellular lysis and its targeting by IpaH7.8
Nat Commun, 14, 2023
|
|
7LNI
 
 | | SeMet CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-05-19 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Clostridioides difficile specific DNA adenine methyltransferase CamA squeezes and flips adenine out of DNA helix.
Nat Commun, 12, 2021
|
|
7LT5
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Cofactor SAH | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-02-18 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Clostridioides difficile specific DNA adenine methyltransferase CamA squeezes and flips adenine out of DNA helix.
Nat Commun, 12, 2021
|
|
7LNJ
 
 | |
5X4R
 
 | | Structure of the N-terminal domain (NTD) of MERS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
7XPQ
 
 | |
7XPP
 
 | |
2P8S
 
 | | Human dipeptidyl peptidase IV/CD26 in complex with a cyclohexalamine inhibitor | | Descriptor: | (1S,2R,5S)-5-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-2-(2,4,5-TRIFLUOROPHENYL)CYCLOHEXANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Biftu, T. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational design of a novel, potent, and orally bioavailable cyclohexylamine DPP-4 inhibitor by application of molecular modeling and X-ray crystallography of sitagliptin
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1F6W
 
 | |
4FYO
 
 | | Crystal structure of spleen tyrosine kinase complexed with N-{(S)-1-[7-(3,4-Dimethoxy-phenylamino)-thiazolo[5,4-d]pyrimidin-5-yl]-pyrrolidin-3-yl}-terephthalamic acid | | Descriptor: | 4-{[(3S)-1-{7-[(3,4-dimethoxyphenyl)amino][1,3]thiazolo[5,4-d]pyrimidin-5-yl}pyrrolidin-3-yl]carbamoyl}benzoic acid, Tyrosine-protein kinase SYK | | Authors: | Kuglstatter, A, Slade, M. | | Deposit date: | 2012-07-05 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rational design of highly selective spleen tyrosine kinase inhibitors.
J.Med.Chem., 55, 2012
|
|
7VQ1
 
 | | Structure of Apo-hsTRPM2 channel | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yu, X.F, Xie, Y, Zhang, X.K, Ma, C, Guo, J.T, Yang, F, Yang, W. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural and functional basis of the selectivity filter as a gate in human TRPM2 channel.
Cell Rep, 37, 2021
|
|
7VQ2
 
 | | Structure of Apo-hsTRPM2 channel TM domain | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yu, X.F, Xie, Y, Zhang, X.K, Ma, C, Guo, J.T, Yang, F, Yang, W. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural and functional basis of the selectivity filter as a gate in human TRPM2 channel.
Cell Rep, 37, 2021
|
|
4FZ6
 
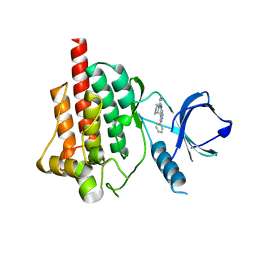 | |
7CBQ
 
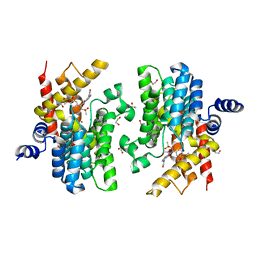 | | Crystal structure of PDE4D catalytic domain in complex with Apremilast | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, N-{2-[(1S)-1-(3-ethoxy-4-methoxyphenyl)-2-(methylsulfonyl)ethyl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}acetamide, ... | | Authors: | Zhang, X.L, Xu, Y.C. | | Deposit date: | 2020-06-13 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Design, synthesis, and biological evaluation of tetrahydroisoquinolines derivatives as novel, selective PDE4 inhibitors for antipsoriasis treatment.
Eur.J.Med.Chem., 211, 2021
|
|
7CHA
 
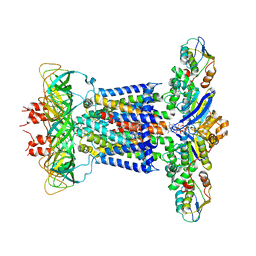 | | Cryo-EM structure of P.aeruginosa MlaFEBD with AMPPNP | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, MlaD domain-containing protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
2MHM
 
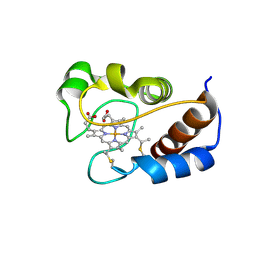 | | Solution structure of cytochrome c Y67H | | Descriptor: | Cytochrome c iso-1, HEME C | | Authors: | Lan, W.X, Wang, Z.H, Yang, Z.Z, Ying, T.L, Wu, H.M, Tan, X.S, Cao, C.Y, Huang, Z.X. | | Deposit date: | 2013-11-26 | | Release date: | 2014-10-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Cytochrome c Y67H Mutant to Function as a Peroxidase
Plos One, 9, 2014
|
|
8H69
 
 | | Cryo-EM structure of influenza RNA polymerase | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*UP*AP*AP*AP*CP*UP*CP*CP*UP*GP*CP*UP*UP*UP*UP*GP*CP*U)-3'), ... | | Authors: | Li, H, Wu, Y, Liang, H, Liu, Y. | | Deposit date: | 2022-10-16 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An intermediate state allows influenza polymerase to switch smoothly between transcription and replication cycles.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7CBJ
 
 | | Crystal structure of PDE4D catalytic domain in complex with compound 36 | | Descriptor: | (1S)-1-[(7-chloranyl-1H-indol-3-yl)methyl]-6,7-dimethoxy-3,4-dihydro-1H-isoquinoline-2-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Xu, Y.C. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design, synthesis, and biological evaluation of tetrahydroisoquinolines derivatives as novel, selective PDE4 inhibitors for antipsoriasis treatment.
Eur.J.Med.Chem., 211, 2021
|
|
4FYN
 
 | |
7CH6
 
 | | Cryo-EM structure of E.coli MlaFEB with AMPPNP | | Descriptor: | Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-08-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
