6LUM
 
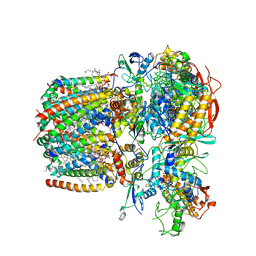 | | Structure of Mycobacterium smegmatis succinate dehydrogenase 2 | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ... | | Authors: | Gao, Y, Gong, H, Zhou, X, Xiao, Y, Wang, W, Ji, W, Wang, Q, Rao, Z. | | Deposit date: | 2020-01-29 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of trimeric Mycobacterium smegmatis succinate dehydrogenase with a membrane-anchor SdhF.
Nat Commun, 11, 2020
|
|
5X5F
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 2 | | Descriptor: | S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
6CXC
 
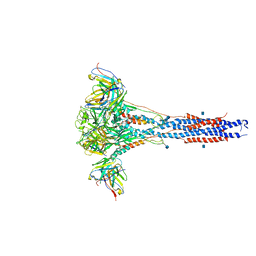 | | 3.9A Cryo-EM structure of murine antibody bound at a novel epitope of respiratory syncytial virus fusion protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, Envelope glycoprotein chimera, ... | | Authors: | Xie, Q, Wang, Z, Chen, X, Ni, F, Ma, J, Wang, Q. | | Deposit date: | 2018-04-02 | | Release date: | 2019-07-31 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure basis of neutralization by a novel site II/IV antibody against respiratory syncytial virus fusion protein.
Plos One, 14, 2019
|
|
7T90
 
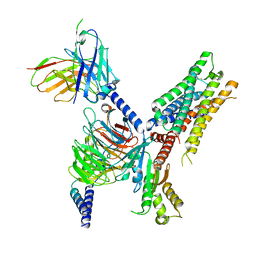 | | Cryo-EM structure of ACh-bound M2R-Go signaling complex in S2 state | | Descriptor: | ACETYLCHOLINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, J, Wang, Q, Du, Y, Kobilka, B.K. | | Deposit date: | 2021-12-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural and dynamic insights into supra-physiological activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nat Commun, 14, 2023
|
|
7T8X
 
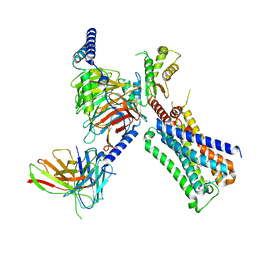 | | Cryo-EM structure of ACh-bound M2R-Go signaling complex in S1 state | | Descriptor: | ACETYLCHOLINE, Antibody fragment, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Wang, Q, Du, Y, Kobilka, B.K. | | Deposit date: | 2021-12-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural and dynamic insights into supra-physiological activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nat Commun, 14, 2023
|
|
7T96
 
 | | Cryo-EM structure of S2 state ACh-bound M2R-Go signaling complex with a PAM | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, ACETYLCHOLINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Wang, Q, Du, Y, Kobilka, B.K. | | Deposit date: | 2021-12-18 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural and dynamic insights into supra-physiological activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nat Commun, 14, 2023
|
|
7T94
 
 | | Cryo-EM structure of S1 state ACh-bound M2R-Go signaling complex with a PAM | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, ACETYLCHOLINE, Antibody fragment, ... | | Authors: | Xu, J, Wang, Q, Du, Y, Kobilka, B.K. | | Deposit date: | 2021-12-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural and dynamic insights into supra-physiological activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nat Commun, 14, 2023
|
|
4HU8
 
 | | Crystal Structure of a Bacterial Ig-like Domain Containing GH10 Xylanase from Termite Gut | | Descriptor: | GH10 Xylanase, GLYCEROL, SULFATE ION | | Authors: | Han, Q, Liu, N, Robinson, H, Cao, L, Qian, C, Wang, Q, Xie, L, Ding, H, Wang, Q, Huang, Y, Li, J, Zhou, Z. | | Deposit date: | 2012-11-02 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical characterization and crystal structure of a GH10 xylanase from termite gut bacteria reveal a novel structural feature and significance of its bacterial Ig-like domain.
Biotechnol.Bioeng., 110, 2013
|
|
9K9U
 
 | | MPXV DNA polymerase complex in editing state 1 | | Descriptor: | DNA (25-MER), DNA (38-MER), DNA polymerase, ... | | Authors: | Xie, Y.F, Kuai, L, Peng, Q, Wang, Q, Wang, H, Li, X.M, Qi, J.X, Ding, Q, Shi, Y, Gao, F. | | Deposit date: | 2024-10-27 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | MPXV DNA polymerase complex in editing state 1
To Be Published
|
|
9K9S
 
 | | MPXV DNA polymerase in complex with primer/4U template DNA | | Descriptor: | DNA (25-MER), DNA (4U 38-MER), DNA polymerase, ... | | Authors: | Xie, Y.F, Kuai, L, Peng, Q, Wang, Q, Wang, H, Li, X.M, Qi, J.X, Ding, Q, Shi, Y, Gao, F. | | Deposit date: | 2024-10-27 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | MPXV DNA polymerase in complex with primer/4U template DNA
To Be Published
|
|
9K9V
 
 | | MPXV DNA polymerase complex in editing state 2 | | Descriptor: | DNA (25-MER), DNA (38-MER), DNA polymerase, ... | | Authors: | Xie, Y.F, Kuai, L, Peng, Q, Wang, Q, Wang, H, Li, X.M, Qi, J.X, Ding, Q, Shi, Y, Gao, F. | | Deposit date: | 2024-10-27 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | MPXV DNA polymerase complex in editing state 2
To Be Published
|
|
9K9R
 
 | | MPXV DNA polymerase in complex with primer/5U template DNA | | Descriptor: | DNA (25-MER), DNA (5U 38-MER), DNA polymerase, ... | | Authors: | Xie, Y.F, Kuai, L, Peng, Q, Wang, Q, Wang, H, Li, X.M, Qi, J.X, Ding, Q, Shi, Y, Gao, F. | | Deposit date: | 2024-10-27 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | MPXV DNA polymerase in complex with primer/5U template DNA
To Be Published
|
|
9K9T
 
 | | MPXV DNA polymerase in complex with CDV | | Descriptor: | DNA (24-MER), DNA (38-MER), DNA polymerase, ... | | Authors: | Xie, Y.F, Kuai, L, Peng, Q, Wang, Q, Wang, H, Li, X.M, Qi, J.X, Ding, Q, Shi, Y, Gao, F. | | Deposit date: | 2024-10-27 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | MPXV DNA polymerase in complex with CDV
To Be Published
|
|
1M8T
 
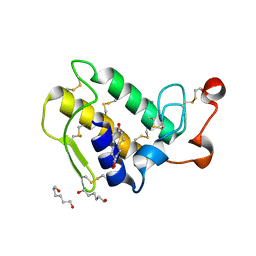 | | Structure of an acidic Phospholipase A2 from the venom of Ophiophagus hannah at 2.1 resolution from a hemihedrally twinned crystal form | | Descriptor: | CALCIUM ION, HEXANE-1,6-DIOL, Phospholipase a2 | | Authors: | Xu, S, Gu, L, Wang, Q, Shu, Y, Lin, Z. | | Deposit date: | 2002-07-26 | | Release date: | 2003-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a king cobra phospholipase A2 determined from a hemihedrally twinned crystal.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3N3Y
 
 | | Crystal structure of Thymidylate Synthase X (ThyX) from Helicobacter pylori with FAD and dUMP at 2.31A resolution | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase thyX | | Authors: | Wang, K, Wang, Q, Chen, J, Chen, L, Jiang, H, Shen, X. | | Deposit date: | 2010-05-21 | | Release date: | 2011-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Structure, Enzymatic Characterization and Inhibitor Discovery of Thymidylate Synthase X (ThyX) from Helicobacter pylori Strain SS1
To be Published
|
|
6PZT
 
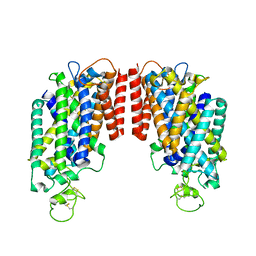 | | cryo-EM structure of human NKCC1 | | Descriptor: | Solute carrier family 12 member 2 | | Authors: | Cao, E, Wang, Q, Yang, X. | | Deposit date: | 2019-08-01 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure of the human cation-chloride cotransporter NKCC1 determined by single-particle electron cryo-microscopy.
Nat Commun, 11, 2020
|
|
4RAZ
 
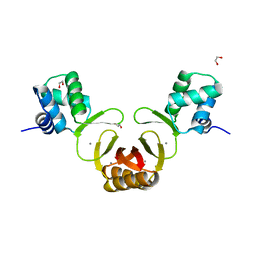 | | Crystal structure of Magnetospirillum gryphiswaldense MSR-1 holo-Fur | | Descriptor: | 1,2-ETHANEDIOL, DNA-binding transcriptional dual regulator of siderophore biosynthesis and transport(Fur family), MANGANESE (II) ION | | Authors: | Deng, Z, Wang, Q, Chen, Z. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator.
Nat Commun, 6
|
|
3DP4
 
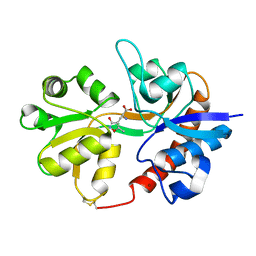 | | Crystal structure of the binding domain of the AMPA subunit GluR3 bound to AMPA | | Descriptor: | (S)-ALPHA-AMINO-3-HYDROXY-5-METHYL-4-ISOXAZOLEPROPIONIC ACID, Glutamate receptor 3, ZINC ION | | Authors: | Ahmed, A.H, Wang, Q, Sondermann, H, Oswald, R.E. | | Deposit date: | 2008-07-07 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of the S1S2 glutamate binding domain of GLuR3.
Proteins, 75, 2008
|
|
3DP6
 
 | | Crystal structure of the binding domain of the AMPA subunit GluR2 bound to glutamate | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Ahmed, A.H, Wang, Q, Sondermann, H, Oswald, R.E. | | Deposit date: | 2008-07-07 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the S1S2 glutamate binding domain of GLuR3.
Proteins, 75, 2008
|
|
3DLN
 
 | | Crystal structure of the binding domain of the AMPA subunit GluR3 bound to glutamate | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 3, ZINC ION | | Authors: | Ahmed, A.H, Wang, Q, Sondermann, H, Oswald, R.E. | | Deposit date: | 2008-06-27 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of the S1S2 glutamate binding domain of GLuR3.
Proteins, 75, 2008
|
|
3FUS
 
 | | Improved Structure of the Unliganded Simian Immunodeficiency Virus gp120 Core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, X, Poon, B, Wang, Q, Ma, J. | | Deposit date: | 2009-01-14 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural improvement of unliganded simian immunodeficiency virus gp120 core by normal-mode-based X-ray crystallographic refinement.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6LNI
 
 | | Cryo-EM structure of amyloid fibril formed by full-length human prion protein | | Descriptor: | Major prion protein | | Authors: | Wang, L.Q, Zhao, K, Yuan, H.Y, Wang, Q, Guan, Z.Y, Tao, J, Li, X.N, Hao, M.M, Chen, J, Zhang, D.L, Zhu, H.L, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2019-12-30 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.702 Å) | | Cite: | Cryo-EM structure of an amyloid fibril formed by full-length human prion protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1QZE
 
 | | HHR23a protein structure based on residual dipolar coupling data | | Descriptor: | UV excision repair protein RAD23 homolog A | | Authors: | Walters, K.J, Lech, P.J, Goh, A.M, Wang, Q, Howley, P.M. | | Deposit date: | 2003-09-16 | | Release date: | 2003-10-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | DNA-repair protein hHR23a alters its protein structure upon binding proteasomal subunit S5a
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1Q6N
 
 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 4 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Protein-tyrosine phosphatase, ... | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
1Q6T
 
 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 11 | | Descriptor: | 6-[4-((2S)-2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-3-{4-[DIFLUORO(PHOSPHONO)METHYL]PHENYL}-2-PHENYLPROPYL)PHENYL]-2-[(1S)-1-METHOXY-3-METHYLBUTYL]QUINOLIN-8-YLPHOSPHONIC ACID, MAGNESIUM ION, Protein-tyrosine phosphatase, ... | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
