7C01
 
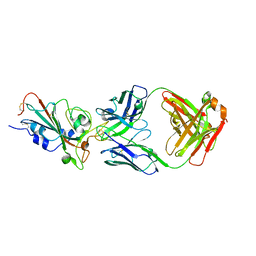 | | Molecular basis for a potent human neutralizing antibody targeting SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CB6 heavy chain, CB6 light chain, ... | | Authors: | Shi, R, Qi, J, Wang, Q, Gao, F.G, Yan, J. | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2.
Nature, 584, 2020
|
|
7TUZ
 
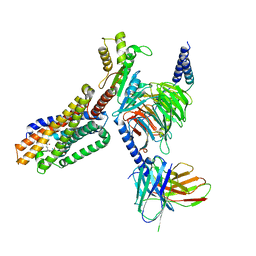 | | Cryo-EM structure of 7alpha,25-dihydroxycholesterol-bound EBI2/GPR183 in complex with Gi protein | | Descriptor: | (2S,4aS,4bS,7R,8S,8aS,9R,10aR)-7-[(2R,3R)-7-hydroxy-3,7-dimethyloctan-2-yl]-4a,7,8-trimethyltetradecahydrophenanthrene-2,9-diol, G-protein coupled receptor 183, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Chen, H, Hung, W, Li, X. | | Deposit date: | 2022-02-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structures of oxysterol sensor EBI2/GPR183, a key regulator of the immune response.
Structure, 30, 2022
|
|
7N9C
 
 | |
7N9E
 
 | |
7N9B
 
 | |
8EKY
 
 | | Cryo-EM structure of the human PRDX4-ErP46 complex | | Descriptor: | Peroxiredoxin-4, Thioredoxin domain-containing protein 5 | | Authors: | Su, C.C. | | Deposit date: | 2022-09-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8EOJ
 
 | | Microsomal triglyceride transfer protein | | Descriptor: | Microsomal triglyceride transfer protein large subunit, Protein disulfide-isomerase | | Authors: | Zhang, Z. | | Deposit date: | 2022-10-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8ENE
 
 | |
8EKW
 
 | | Cryo-EM structure of human PRDX4 | | Descriptor: | Peroxiredoxin-4 | | Authors: | Su, C.C. | | Deposit date: | 2022-09-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8EOR
 
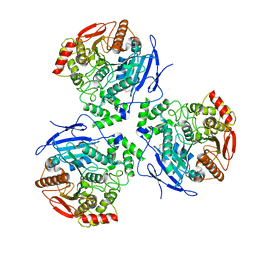 | | Liver carboxylesterase 1 | | Descriptor: | ETHYL ACETATE, Liver carboxylesterase 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zhang, Z, Yu, E. | | Deposit date: | 2022-10-04 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8EM2
 
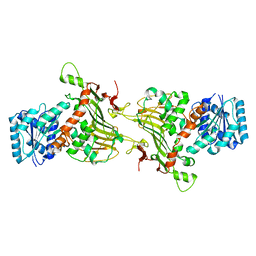 | |
8FEX
 
 | |
4TZR
 
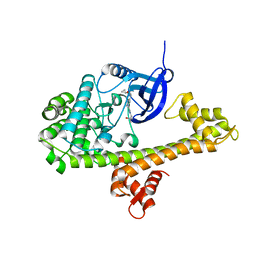 | | Calcium-Dependent Protein Kinase 1 from Toxoplasma gondii (TgCDPK1) in complex with inhibitor UW1561 | | Descriptor: | 1-{4-amino-3-[2-(cyclopropyloxy)quinolin-6-yl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}-2-methylpropan-2-ol, Calmodulin-domain protein kinase 1 | | Authors: | Merritt, E.A. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of an Orally Available and Central Nervous System (CNS) Penetrant Toxoplasma gondii Calcium-Dependent Protein Kinase 1 (TgCDPK1) Inhibitor with Minimal Human Ether-a-go-go-Related Gene (hERG) Activity for the Treatment of Toxoplasmosis.
J. Med. Chem., 59, 2016
|
|
7N9A
 
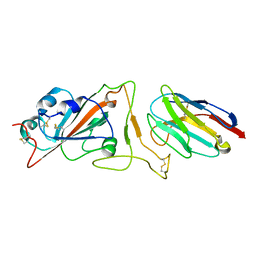 | |
8FFI
 
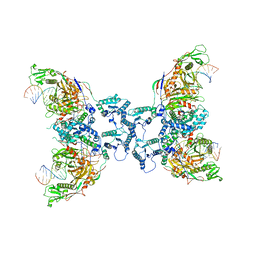 | | Structure of tetramerized MapSPARTA upon guide RNA-mediated target DNA binding | | Descriptor: | MAGNESIUM ION, TIR-APAZ, guide RNA, ... | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Oligomerization-mediated activation of a short prokaryotic Argonaute.
Nature, 621, 2023
|
|
8OU0
 
 | |
8OTZ
 
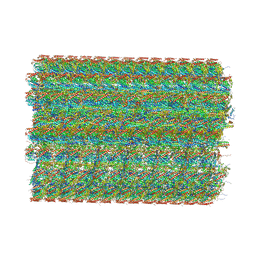 | | 48-nm repeat of the native axonemal doublet microtubule from bovine sperm | | Descriptor: | ATP6V1F neighbor, CFAP97 domain containing 1, Chromosome 13 C20orf85 homolog, ... | | Authors: | Leung, M.R, Zeng, J, Zhang, R, Zeev-Ben-Mordehai, T. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural specializations of the sperm tail.
Cell, 186, 2023
|
|
2H60
 
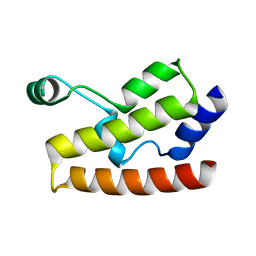 | | Solution Structure of Human Brg1 Bromodomain | | Descriptor: | Probable global transcription activator SNF2L4 | | Authors: | Shen, W, Xu, C, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2006-05-30 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails
Biochemistry, 46, 2007
|
|
1BS1
 
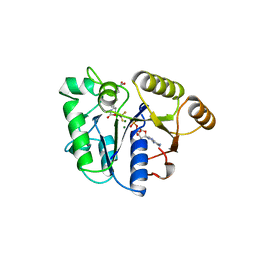 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH DETHIOBIOTIN, ADP , INORGANIC PHOSPHATE AND MAGNESIUM | | Descriptor: | 8-AMINO-7-CARBOXYAMINO-NONANOIC ACID WITH ALUMINUM FLUORIDE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kaeck, H, Sandmark, J, Gibson, K.J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1998-08-31 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of two quaternary complexes of dethiobiotin synthetase, enzyme-MgADP-AlF3-diaminopelargonic acid and enzyme-MgADP-dethiobiotin-phosphate; implications for catalysis.
Protein Sci., 7, 1998
|
|
8HUG
 
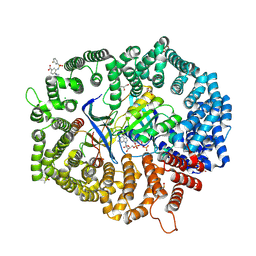 | | F1 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 4-[4-(3-chlorophenyl)piperazin-1-yl]-3-[(3-fluorophenyl)sulfonylamino]benzoic acid, CHLORIDE ION, CRM1 isoform 1, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2022-12-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a Hidden Pocket beneath the NES Groove by Novel Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
8HUF
 
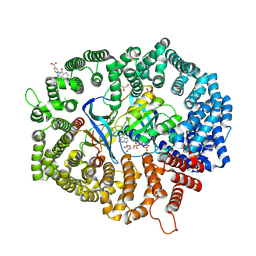 | | B28 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 3-[(4-chlorophenyl)carbonylamino]-4-[4-(2,5-dimethylphenyl)piperazin-1-yl]benzoic acid, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2022-12-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of a Hidden Pocket beneath the NES Groove by Novel Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
1PZV
 
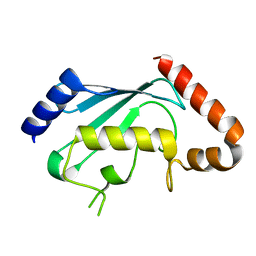 | | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans | | Descriptor: | Probable ubiquitin-conjugating enzyme E2-19 kDa | | Authors: | Schormann, N, Lin, G, Li, S, Symersky, J, Qiu, S, Finley, J, Luo, D, Stanton, A, Carson, M, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-07-14 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans
To be Published
|
|
1A0Q
 
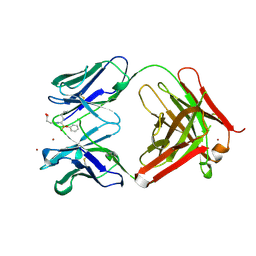 | | 29G11 COMPLEXED WITH PHENYL [1-(1-N-SUCCINYLAMINO)PENTYL] PHOSPHONATE | | Descriptor: | 29G11 FAB (HEAVY CHAIN), 29G11 FAB (LIGHT CHAIN), PHENYL[1-(N-SUCCINYLAMINO)PENTYL]PHOSPHONATE, ... | | Authors: | Buchbinder, J.L, Stephenson, R.C, Scanlan, T.S, Fletterick, R.J. | | Deposit date: | 1997-12-05 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A comparison of the crystallographic structures of two catalytic antibodies with esterase activity.
J.Mol.Biol., 282, 1998
|
|
1BYI
 
 | | STRUCTURE OF APO-DETHIOBIOTIN SYNTHASE AT 0.97 ANGSTROMS RESOLUTION | | Descriptor: | DETHIOBIOTIN SYNTHASE | | Authors: | Sandalova, T, Schneider, G, Kaeck, H, Lindqvist, Y. | | Deposit date: | 1998-10-15 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Structure of dethiobiotin synthetase at 0.97 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6BE1
 
 | | Cryo-EM structure of serotonin receptor | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Basak, S, Chakrapani, S. | | Deposit date: | 2017-10-24 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Cryo-EM structure of 5-HT
Nat Commun, 9, 2018
|
|
