2H2G
 
 | |
2H2I
 
 | | The Structural basis of Sirtuin Substrate Affinity | | Descriptor: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, NAD-dependent deacetylase, ZINC ION | | Authors: | Cosgrove, M.S, Wolberger, C. | | Deposit date: | 2006-05-18 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of sirtuin substrate affinity
Biochemistry, 45, 2006
|
|
2N7S
 
 | | Solution Structure of Leptospiral LigA4 Big Domain | | Descriptor: | Ig-like repeat domain protein 1 | | Authors: | Mei, S. | | Deposit date: | 2015-09-17 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of leptospiral LigA4 Big domain.
Biochem. Biophys. Res. Commun., 467, 2015
|
|
2NB4
 
 | | Solution structure of Q388A3 PDZ domain | | Descriptor: | Putative uncharacterized protein | | Authors: | Mei, S. | | Deposit date: | 2016-01-24 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Q388A3 PDZ domain from Trypanosoma brucei
J.Struct.Biol., 194, 2016
|
|
3W94
 
 | |
7BX8
 
 | | Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in symmetric "resting state" | | Descriptor: | Integral membrane indolylacetylinositol arabinosyltransferase EmbB, Meromycolate extension acyl carrier protein | | Authors: | Gao, R.G, Zhang, L, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-04-17 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB2-AcpM2.
Protein Cell, 11, 2020
|
|
7E1F
 
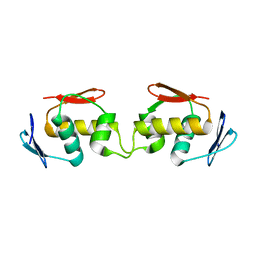 | | Native-DBD | | Descriptor: | DNA-binding response regulator | | Authors: | Hong, S, Zhang, P. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.447 Å) | | Cite: | Structural basis of phosphorylation-induced activation of the response regulator VbrR.
Acta Biochim.Biophys.Sin., 2023
|
|
7E1D
 
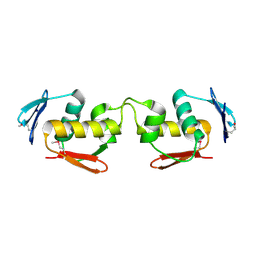 | | Se-DBD | | Descriptor: | DNA-binding response regulator | | Authors: | Hong, S, Zhang, P. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-09 | | Last modified: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structural basis of phosphorylation-induced activation of the response regulator VbrR.
Acta Biochim.Biophys.Sin., 2023
|
|
5IKL
 
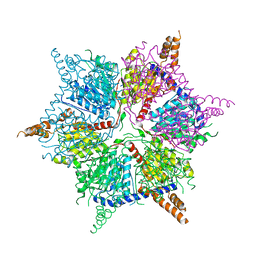 | |
2FHS
 
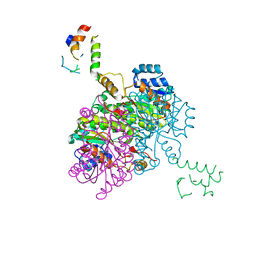 | | Structure of Acyl Carrier Protein Bound to FabI, the Enoyl Reductase from Escherichia Coli | | Descriptor: | Acyl carrier protein, enoyl-[acyl-carrier-protein] reductase, NADH-dependent | | Authors: | Kolappan, S, Novichenok, P, Rafi, S, Simmerling, C, Tonge, P.J, Kisker, C. | | Deposit date: | 2005-12-27 | | Release date: | 2006-10-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Acyl Carrier Protein Bound to FabI, the FASII Enoyl Reductase from Escherichia coli.
J.Biol.Chem., 281, 2006
|
|
3HDB
 
 | | Crystal structure of AaHIV, A metalloproteinase from venom of Agkistrodon Acutus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AAHIV, CALCIUM ION, ... | | Authors: | Zhu, Z.Q, Niu, L.W, Teng, M.K. | | Deposit date: | 2009-05-07 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis of the autolysis of AaHIV suggests a novel target recognizing model for ADAM/reprolysin family proteins
Biochem.Biophys.Res.Commun., 386, 2009
|
|
2JUS
 
 | |
2MQG
 
 | |
2JSZ
 
 | | Solution structure of Tpx in the reduced state | | Descriptor: | Probable thiol peroxidase | | Authors: | Jin, C, Lu, J, Yang, F. | | Deposit date: | 2007-07-17 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Reversible conformational switch revealed by the redox structures of Bacillus subtilis thiol peroxidase
Biochem.Biophys.Res.Commun., 373, 2008
|
|
6IWI
 
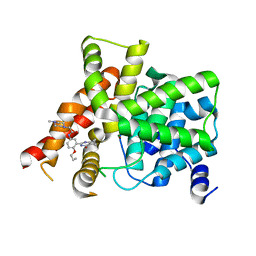 | | Crystal structure of PDE5A in complex with a novel inhibitor | | Descriptor: | MAGNESIUM ION, N-[3-(4,5-diethyl-6-oxo-1,6-dihydropyrimidin-2-yl)-4-propoxyphenyl]-2-(4-methylpiperazin-1-yl)acetamide, ZINC ION, ... | | Authors: | Zhang, X.L, Xu, Y.C. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Pharmacokinetics-Driven Optimization of 4(3 H)-Pyrimidinones as Phosphodiesterase Type 5 Inhibitors Leading to TPN171, a Clinical Candidate for the Treatment of Pulmonary Arterial Hypertension.
J.Med.Chem., 62, 2019
|
|
2LI6
 
 | |
1OO4
 
 | |
2JUT
 
 | |
2H2D
 
 | |
2H2F
 
 | |
2JSY
 
 | | Solution structure of Tpx in the oxidized state | | Descriptor: | Probable thiol peroxidase | | Authors: | Jin, C, Lu, J. | | Deposit date: | 2007-07-17 | | Release date: | 2008-07-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Reversible conformational switch revealed by the redox structures of Bacillus subtilis thiol peroxidase
Biochem.Biophys.Res.Commun., 373, 2008
|
|
2JUR
 
 | |
2JUQ
 
 | |
2MNY
 
 | | NMR Structure of KDM5B PHD1 finger | | Descriptor: | Lysine-specific demethylase 5B, ZINC ION | | Authors: | Zhang, Y, Yang, H.R, Guo, X, Rong, N.Y, Song, Y.J, Xu, Y.W, Lan, W.X, Xu, Y.H, Cao, C. | | Deposit date: | 2014-04-16 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The PHD1 finger of KDM5B recognizes unmodified H3K4 during the demethylation of histone H3K4me2/3 by KDM5B.
Protein Cell, 5, 2014
|
|
2FDW
 
 | | Crystal Structure Of Human Microsomal P450 2A6 with the inhibitor (5-(Pyridin-3-yl)furan-2-yl)methanamine bound | | Descriptor: | (5-(PYRIDIN-3-YL)FURAN-2-YL)METHANAMINE, Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yano, J.K, Stout, C.D, Johnson, E.F. | | Deposit date: | 2005-12-14 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthetic Inhibitors of Cytochrome P-450 2A6: Inhibitory Activity, Difference Spectra, Mechanism of Inhibition, and Protein Cocrystallization.
J.Med.Chem., 49, 2006
|
|
