4WSZ
 
 | |
4WT0
 
 | |
4WUH
 
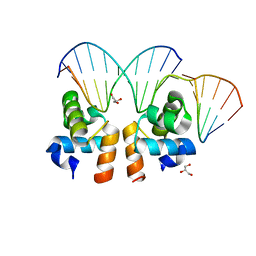 | | Crystal structure of E. faecalis DNA binding domain LiaR wild type complexed with 22bp DNA | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*CP*G)-3'), DNA (5'-D(P*GP*GP*AP*CP*TP*TP*AP*AP*GP*AP*AP*CP*GP*AP*TP*TP*T)-3'), DNA (5'-D(P*TP*TP*CP*TP*TP*AP*AP*GP*TP*CP*C)-3'), ... | | Authors: | Davlieva, M, Shamoo, Y. | | Deposit date: | 2014-10-31 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | A variable DNA recognition site organization establishes the LiaR-mediated cell envelope stress response of enterococci to daptomycin.
Nucleic Acids Res., 43, 2015
|
|
4WU4
 
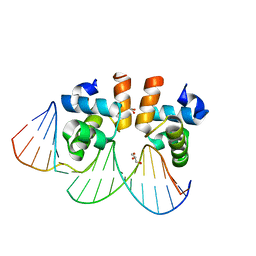 | | Crystal structure of E. faecalis DNA binding domain LiaRD191N complexed with 22bp DNA | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*CP*GP*TP*TP*CP*TP*TP*AP*AP*GP*TP*CP*C)-3'), DNA (5'-D(P*GP*GP*AP*CP*TP*TP*AP*AP*GP*AP*AP*CP*GP*AP*TP*TP*T)-3'), GLYCEROL, ... | | Authors: | Davlieva, M, Shamoo, Y. | | Deposit date: | 2014-10-31 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A variable DNA recognition site organization establishes the LiaR-mediated cell envelope stress response of enterococci to daptomycin.
Nucleic Acids Res., 43, 2015
|
|
4WUL
 
 | |
7N85
 
 | | Inner ring spoke from the isolated yeast NPC | | Descriptor: | Nucleoporin ASM4, Nucleoporin NIC96, Nucleoporin NSP1, ... | | Authors: | Akey, C.W, Rout, M.P, Ouch, C, Echevarria, I, Fernandez-Martinez, J, Nudelman, I. | | Deposit date: | 2021-06-13 | | Release date: | 2022-01-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
7N84
 
 | | Double nuclear outer ring from the isolated yeast NPC | | Descriptor: | Nucleoporin 145c, Nucleoporin NUP120, Nucleoporin NUP133, ... | | Authors: | Akey, C.W, Rout, M.P, Ouch, C, Echevarria, I, Fernandez-Martinez, J, Nudelman, I. | | Deposit date: | 2021-06-13 | | Release date: | 2022-01-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
7N9F
 
 | | Structure of the in situ yeast NPC | | Descriptor: | Dynein light chain 1, cytoplasmic, Nucleoporin 145c, ... | | Authors: | Villa, E, Singh, D, Ludtke, S.J, Akey, C.W, Rout, M.P, Echeverria, I, Suslov, S. | | Deposit date: | 2021-06-17 | | Release date: | 2022-01-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (37 Å) | | Cite: | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
3SOW
 
 | |
8J5P
 
 | | Cryo-EM structure of native RC-LH complex from Roseiflexus castenholzii at 2,000lux | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, ... | | Authors: | Xu, X, Xin, J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Carotenoid assembly regulates quinone diffusion and the Roseiflexus castenholzii reaction center-light harvesting complex architecture.
Elife, 12, 2023
|
|
8J5O
 
 | | Cryo-EM structure of native RC-LH complex from Roseiflexus castenholzii at 100lux | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, ... | | Authors: | Xu, X, Xin, J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Carotenoid assembly regulates quinone diffusion and the Roseiflexus castenholzii reaction center-light harvesting complex architecture.
Elife, 12, 2023
|
|
8K8E
 
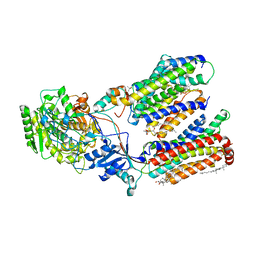 | | Human gamma-secretase in complex with a substrate mimetic | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shi, Y.G, Zhou, R, Wolfe, M.S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Familial Alzheimer mutations stabilize synaptotoxic gamma-secretase-substrate complexes.
Cell Rep, 43, 2024
|
|
7PZR
 
 | | Cryo-EM structure of POLRMT in free form. | | Descriptor: | DNA-directed RNA polymerase, mitochondrial | | Authors: | Das, H, Hallberg, B.M. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Non-coding 7S RNA inhibits transcription via mitochondrial RNA polymerase dimerization.
Cell, 185, 2022
|
|
3SOU
 
 | |
3SOX
 
 | | Structure of UHRF1 PHD finger in the free form | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Rajakumara, E, Patel, D.J. | | Deposit date: | 2011-06-30 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6501 Å) | | Cite: | PHD Finger Recognition of Unmodified Histone H3R2 Links UHRF1 to Regulation of Euchromatic Gene Expression.
Mol.Cell, 43, 2011
|
|
5H3X
 
 | |
6CF0
 
 | | Sperm Whale Myoglobin H64V Mutant with Nitrite | | Descriptor: | GLYCEROL, Myoglobin, NITRITE ION, ... | | Authors: | Powell, S.M, Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2018-02-13 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Nitrosyl Myoglobins and Their Nitrite Precursors: Crystal Structural and Quantum Mechanics and Molecular Mechanics Theoretical Investigations of Preferred Fe -NO Ligand Orientations in Myoglobin Distal Pockets.
Biochemistry, 57, 2018
|
|
5H3W
 
 | |
3QT0
 
 | | Revealing a steroid receptor ligand as a unique PPARgamma agonist | | Descriptor: | 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, Nuclear receptor coactivator 1 peptide, Peroxisome proliferator-activated receptor gamma | | Authors: | Rong, H. | | Deposit date: | 2011-02-22 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | Revealing a steroid receptor ligand as a unique PPAR gamma agonist.
Cell Res., 22, 2012
|
|
8HJV
 
 | | Cryo-EM structure of carotenoid-depleted RC-LH complex from Roseiflexus castenholzii at 10,000 lux | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, ... | | Authors: | Xu, X, Xin, J. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Carotenoid assembly regulates quinone diffusion and the Roseiflexus castenholzii reaction center-light harvesting complex architecture.
Elife, 12, 2023
|
|
8HJU
 
 | | Cryo-EM structure of native RC-LH complex from Roseiflexus castenholzii at 10,000 lux | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, ... | | Authors: | Xu, X, Xin, J. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Carotenoid assembly regulates quinone diffusion and the Roseiflexus castenholzii reaction center-light harvesting complex architecture.
Elife, 12, 2023
|
|
7CVA
 
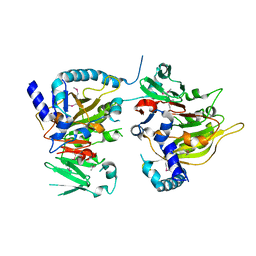 | | RNA methyltransferase METTL4 | | Descriptor: | Methyltransferase-like protein 2 | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CV9
 
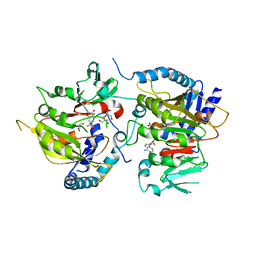 | | RNA methyltransferase METTL4 | | Descriptor: | GLYCEROL, Methyltransferase-like protein 2, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CV6
 
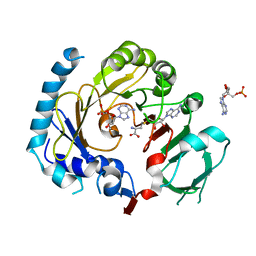 | | RNA methyltransferase METTL4 | | Descriptor: | 2'-O-methyladenosine 5'-(dihydrogen phosphate), Methyltransferase-like protein 2, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CV7
 
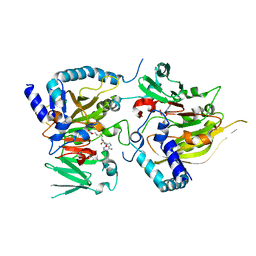 | | RNA methyltransferase METTL4 | | Descriptor: | Methyltransferase-like protein 2, S-ADENOSYLMETHIONINE | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
