3PB1
 
 | | Crystal Structure of a Michaelis Complex between Plasminogen Activator Inhibitor-1 and Urokinase-type Plasminogen Activator | | Descriptor: | Plasminogen activator inhibitor 1, Plasminogen activator, urokinase, ... | | Authors: | Lin, Z, Jiang, L, Huang, M, Structure 2 Function Project (S2F) | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition of urokinase-type plasminogen activator by plasminogen activator inhibitor-1.
J.Biol.Chem., 286, 2011
|
|
4R28
 
 | |
3RDH
 
 | | X-ray induced covalent inhibition of 14-3-3 | | Descriptor: | 14-3-3 protein zeta/delta, 4-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]benzoic acid, NICKEL (II) ION | | Authors: | Horton, J.R, Upadhyay, A.K, Fu, H, Cheng, X. | | Deposit date: | 2011-04-01 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery and structural characterization of a small molecule 14-3-3 protein-protein interaction inhibitor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7DRT
 
 | | Human Wntless in complex with Wnt3a | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhong, Q, Zhao, Y, Ye, F, Xiao, Z, Huang, G, Zhang, Y, Lu, P, Xu, W, Zhou, Q, Ma, D. | | Deposit date: | 2020-12-29 | | Release date: | 2021-07-14 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM structure of human Wntless in complex with Wnt3a.
Nat Commun, 12, 2021
|
|
3NBW
 
 | | X-ray structure of ketohexokinase in complex with a pyrazole compound | | Descriptor: | 5-amino-3-(methylsulfanyl)-1-phenyl-1H-pyrazole-4-carbonitrile, GLYCEROL, Ketohexokinase, ... | | Authors: | Abad, M.C, Gibbs, A.C, Spurlino, J.C. | | Deposit date: | 2010-06-04 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Electron density guided fragment-based lead discovery of ketohexokinase inhibitors.
J.Med.Chem., 53, 2010
|
|
3NC2
 
 | |
3NC9
 
 | |
3NMU
 
 | | Crystal Structure of substrate-bound halfmer box C/D RNP | | Descriptor: | 50S ribosomal protein L7Ae, Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, NOP5/NOP56 related protein, ... | | Authors: | Li, H, Xue, S, Wang, R. | | Deposit date: | 2010-06-22 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.729 Å) | | Cite: | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle.
Mol.Cell, 39, 2010
|
|
3NCA
 
 | | X-ray structure of ketohexokinase in complex with a thieno pyridinol compound | | Descriptor: | Ketohexokinase, SULFATE ION, thieno[3,2-b]pyridin-7-ol | | Authors: | Abad, M.C, Gibbs, A.C, Spurlino, J.C. | | Deposit date: | 2010-06-04 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Electron density guided fragment-based lead discovery of ketohexokinase inhibitors.
J.Med.Chem., 53, 2010
|
|
3QE7
 
 | | Crystal Structure of Uracil Transporter--UraA | | Descriptor: | URACIL, Uracil permease, nonyl beta-D-glucopyranoside | | Authors: | Lu, F.R, Li, S, Yan, N. | | Deposit date: | 2011-01-20 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.781 Å) | | Cite: | Structure and mechanism of the uracil transporter UraA
Nature, 472, 2011
|
|
3HIE
 
 | |
4PBA
 
 | |
2L83
 
 | | A protein from Haloferax volcanii | | Descriptor: | Small archaeal modifier protein 1 | | Authors: | Zhang, W, Liao, S, Fan, K, Tu, X. | | Deposit date: | 2011-01-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ionic strength-dependent conformations of a ubiquitin-like small archaeal modifier protein (SAMP1) from Haloferax volcanii.
Protein Sci., 22, 2013
|
|
5X4R
 
 | | Structure of the N-terminal domain (NTD) of MERS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
7YFE
 
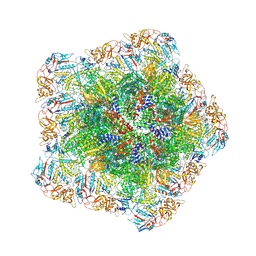 | | In situ structure of polymerase complex of mammalian reovirus in virion | | Descriptor: | Lambda-2 protein, Mu-2 protein, RNA (5'-R(P*AP*CP*GP*AP*UP*UP*AP*GP*C)-3'), ... | | Authors: | Bao, K.Y, Zhang, X.L, Li, D.Y, Zhu, P. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YED
 
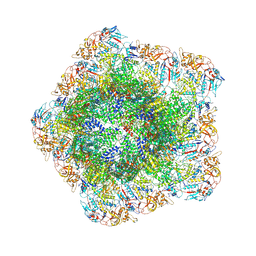 | | In situ structure of polymerase complex of mammalian reovirus in the elongation state | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Lambda-2 protein, MAGNESIUM ION, ... | | Authors: | Bao, K.Y, Zhang, X.L, Li, D.Y, Zhu, P. | | Deposit date: | 2022-07-05 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4Q9V
 
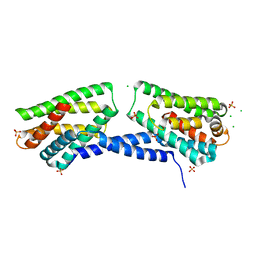 | | Crystal structure of TIPE3 | | Descriptor: | CHLORIDE ION, SULFATE ION, Tumor necrosis factor alpha-induced protein 8-like protein 3 | | Authors: | Wu, J, Zhang, X, Chen, Y.H, Shi, Y. | | Deposit date: | 2014-05-02 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | TIPE3 Is the Transfer Protein of Lipid Second Messengers that Promote Cancer.
Cancer Cell, 26, 2014
|
|
6IPQ
 
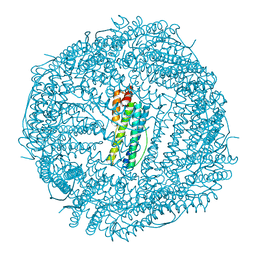 | | Non-native ferritin 8-mer mutant-C90A/C102A/C130A | | Descriptor: | Ferritin heavy chain, MAGNESIUM ION | | Authors: | Zang, J, Chen, H, Wang, Y, Zhao, G. | | Deposit date: | 2018-11-03 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Disulfide-mediated conversion of 8-mer bowl-like protein architecture into three different nanocages.
Nat Commun, 10, 2019
|
|
7RCU
 
 | | Synthetic Max homodimer mimic in complex with DNA | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 2-(2,5-dioxopyrrolidin-1-yl)acetamide, ACETAMIDE, ... | | Authors: | Speltz, T, Qiao, Z, Shangguan, S, Fanning, S, Greene, J, Moellering, R. | | Deposit date: | 2021-07-08 | | Release date: | 2022-09-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Targeting MYC with modular synthetic transcriptional repressors derived from bHLH DNA-binding domains.
Nat.Biotechnol., 41, 2023
|
|
5H05
 
 | | Crystal structure of AmyP E221Q in complex with MALTOTRIOSE | | Descriptor: | AmyP, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | He, C, Liu, Y. | | Deposit date: | 2016-10-03 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of a raw-starch-degrading bacterial alpha-amylase belonging to subfamily 37 of the glycoside hydrolase family GH13
Sci Rep, 7, 2017
|
|
2JSZ
 
 | | Solution structure of Tpx in the reduced state | | Descriptor: | Probable thiol peroxidase | | Authors: | Jin, C, Lu, J, Yang, F. | | Deposit date: | 2007-07-17 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Reversible conformational switch revealed by the redox structures of Bacillus subtilis thiol peroxidase
Biochem.Biophys.Res.Commun., 373, 2008
|
|
8GYD
 
 | | Structure of Schistosoma japonicum Glutathione S-transferase bound with the ligand complex of 16 | | Descriptor: | (2R)-2-[[2-(5-chloranylthiophen-2-yl)-4-oxidanylidene-6-[2-(1H-1,2,3,4-tetrazol-5-yl)phenyl]quinazolin-3-yl]methyl]-3-(4-chlorophenyl)propanoic acid, ETHANOL, Glutathione S-transferase class-mu 26 kDa isozyme | | Authors: | Wen, X, Jin, R, Hu, H, Zhu, J, Song, W, Lu, X. | | Deposit date: | 2022-09-22 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery, SAR Study of GST Inhibitors from a Novel Quinazolin-4(1 H )-one Focused DNA-Encoded Library.
J.Med.Chem., 66, 2023
|
|
5X4S
 
 | | Structure of the N-terminal domain (NTD)of SARS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
2JUT
 
 | |
6AKK
 
 | | Crystal structure of the second Coiled-coil domain of SIKE1 | | Descriptor: | GLYCEROL, Suppressor of IKBKE 1 | | Authors: | Zhou, L, Chen, M, Zhou, Z.C. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
