3U1S
 
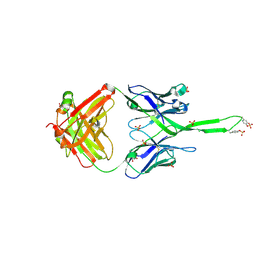 | | Crystal structure of human Fab PGT145, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | Fab PGT145 Heavy chain, Fab PGT145 Light chain, GLYCEROL, ... | | Authors: | Julien, J.-P, Diwanji, D, Burton, D.R, Wilson, I.A. | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
2HAV
 
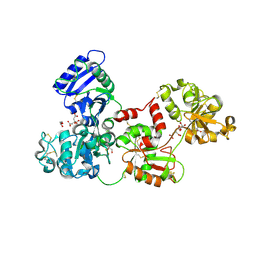 | | Apo-Human Serum Transferrin (Glycosylated) | | Descriptor: | CITRIC ACID, GLYCEROL, Serotransferrin | | Authors: | Wally, J, Everse, S.J. | | Deposit date: | 2006-06-13 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of Iron-free Human Serum Transferrin Provides Insight into Inter-lobe Communication and Receptor Binding.
J.Biol.Chem., 281, 2006
|
|
3QD5
 
 | |
2KL5
 
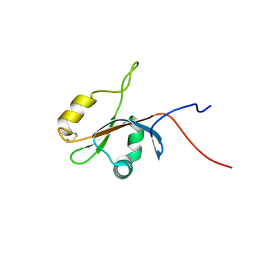 | | Solution NMR Structure of protein yutD from B.subtilis, Northeast Structural Genomics Consortium Target SR232 | | Descriptor: | Uncharacterized protein yutD | | Authors: | Liu, G, Hamilton, K, Xiao, R, Ciccosanti, C, Ho, C.J, Everett, J, Nair, R, Acton, T, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of protein yutD from B.subtilis, Northeast Structural Genomics Consortium Target Target SR232
To be Published
|
|
3TWD
 
 | | glmuC1 in complex with an antibacterial inhibitor | | Descriptor: | 4-({5-[(4-aminophenyl)(phenyl)sulfamoyl]-2,4-dimethoxyphenyl}amino)-4-oxobutanoic acid, Bifunctional protein glmU, SULFATE ION | | Authors: | Lahiri, S, Otterbein, L. | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | In Vitro Validation of Acetyltransferase Activity of GlmU as an Antibacterial Target in Haemophilus influenzae.
J.Biol.Chem., 286, 2011
|
|
3U4B
 
 | | CH04H/CH02L Fab P4 | | Descriptor: | CH02 Light chain, CH04 Heavy chain | | Authors: | Pancera, M, Louder, R, Mclellan, J.S, KWong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3U36
 
 | | Crystal Structure of PG9 Fab | | Descriptor: | PG9 Fab heavy chain, PG9 Fab light chain, SULFATE ION | | Authors: | McLellan, J.S, Kwong, P.D. | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.281 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
2KT9
 
 | | Solution NMR Structure of Probable 30S Ribosomal Protein PSRP-3 (Ycf65-like protein) from Synechocystis sp. (strain PCC 6803), Northeast Structural Genomics Consortium Target Target SgR46 | | Descriptor: | Probable 30S ribosomal protein PSRP-3 | | Authors: | Liu, G, Janjua, J, Xiao, R, Mao, B, Buchwald, W.A, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Probable 30S ribosomal protein PSRP-3 (Ycf65-like protein) from Synechocystis sp. (PCC 6803), Northeast Structural Genomics Consortium Target Target SgR46
To be Published
|
|
2KXV
 
 | |
3QPF
 
 | | Analysis of a New Family of Widely Distributed Metal-independent alpha-Mannosidases Provides Unique Insight into the Processing of N-linked Glycans, Streptococcus pneumoniae SP_2144 apo-structure | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein | | Authors: | Gregg, K.J, Zandberg, W.F, Hehemann, J.-H, Whitworth, G.E, Deng, L.E, Vocadlo, D.J. | | Deposit date: | 2011-02-12 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Analysis of a New Family of Widely Distributed Metal-independent {alpha}-Mannosidases Provides Unique Insight into the Processing of N-Linked Glycans.
J.Biol.Chem., 286, 2011
|
|
3QH2
 
 | | Crystal structure of TenI from Bacillus subtilis complexed with product cThz-P | | Descriptor: | 4-methyl-5-[2-(phosphonooxy)ethyl]-1,3-thiazole-2-carboxylic acid, Regulatory protein tenI, SULFATE ION | | Authors: | Han, Y, Zhang, Y, Hazra, A, Chatterjee, A, Lai, R, Begley, T.P, Ealick, S.E. | | Deposit date: | 2011-01-25 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | A Missing Enzyme in Thiamin Thiazole Biosynthesis: Identification of TenI as a Thiazole Tautomerase.
J.Am.Chem.Soc., 133, 2011
|
|
2KXT
 
 | |
2FFZ
 
 | |
3TWC
 
 | |
3TYG
 
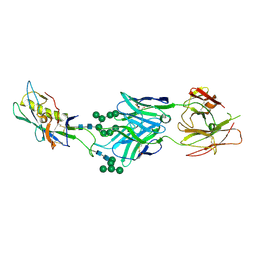 | | Crystal structure of broad and potent HIV-1 neutralizing antibody PGT128 in complex with a glycosylated engineered gp120 outer domain with miniV3 (eODmV3) | | Descriptor: | Envelope glycoprotein gp160, PGT128 heavy chain, Ig gamma-1 chain C region, ... | | Authors: | Pejchal, R, Huang, P.S, Schief, W.R, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2011-09-25 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield.
Science, 334, 2011
|
|
3U4E
 
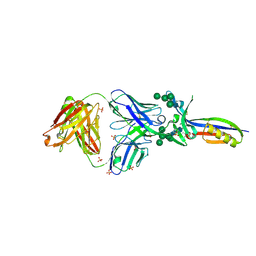 | | Crystal Structure of PG9 Fab in Complex with V1V2 Region from HIV-1 strain CAP45 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PG9 Heavy Chain, PG9 Light Chain, ... | | Authors: | Gorman, J, McLellan, J, Pancera, M, Kwong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3SGM
 
 | | Bromoderivative-2 of amyloid-related segment of alphaB-crystallin residues 90-100 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha-crystallin B chain | | Authors: | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2011-06-15 | | Release date: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (1.7006 Å) | | Cite: | Atomic view of a toxic amyloid small oligomer.
Science, 335, 2012
|
|
3SGR
 
 | | Tandem repeat of amyloid-related segment of alphaB-crystallin residues 90-100 mutant V91L | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Tandem repeat of amyloid-related segment of alphaB-crystallin residues 90-100 mutant V91L | | Authors: | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2011-06-15 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Atomic view of a toxic amyloid small oligomer.
Science, 335, 2012
|
|
3U2S
 
 | | Crystal Structure of PG9 Fab in Complex with V1V2 Region from HIV-1 strain ZM109 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | McLellan, J.S, Pancera, M, Kwong, P.D. | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
2KRK
 
 | | Solution NMR Structure of 26S protease regulatory subunit 8 from H.sapiens, Northeast Structural Genomics Consortium Target Target HR3102A | | Descriptor: | 26S protease regulatory subunit 8 | | Authors: | Liu, G, Janjua, J, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of 26S protease regulatory subunit 8 from H.sapiens, Northeast Structural Genomics Consortium Target HR3102A
To be Published
|
|
2JEW
 
 | |
3U46
 
 | | CH04H/CH02L P212121 | | Descriptor: | CH02 Light chain Fab, CH04 Heavy chain Fab | | Authors: | Louder, R, Pancera, M, McLellan, J.S, Kwong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
2LQ1
 
 | | Solution structure of de novo designed antifreeze peptide 3 | | Descriptor: | de novo designed antifreeze peptide 3 | | Authors: | Bhunia, A. | | Deposit date: | 2012-02-21 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures, dynamics, and ice growth inhibitory activity of Peptide fragments derived from an antarctic yeast protein
Plos One, 7, 2012
|
|
2L2G
 
 | | Solution structure of Opossum Domain 11 | | Descriptor: | IGF2R DOMAIN 11 | | Authors: | Williams, C, Hoppe, H, Rezgui, D, Rezgui, M, Frago, S, Ellis, R.Z, Wattana-Amorn, P, Prince, S.N, Zaccheo, O.J, Forbes, B, Jones, E.Y, Crump, M.P, Bassim, A.H. | | Deposit date: | 2010-08-18 | | Release date: | 2012-02-15 | | Last modified: | 2015-02-25 | | Method: | SOLUTION NMR | | Cite: | An exon splice enhancer primes IGF2:IGF2R binding site structure and function evolution.
Science, 338, 2012
|
|
2LQ0
 
 | | Solution structure of de novo designed antifreeze peptide 1m | | Descriptor: | de novo designed antifreeze peptide 1m | | Authors: | Bhunia, A. | | Deposit date: | 2012-02-21 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures, dynamics, and ice growth inhibitory activity of Peptide fragments derived from an antarctic yeast protein
Plos One, 7, 2012
|
|
