7PQS
 
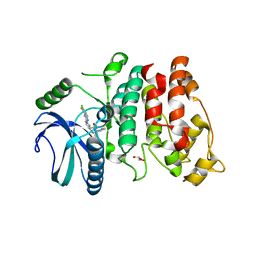 | | SRPK1 in complex with MSC2711186 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Schroeder, M, Leiendecker, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-09-20 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | SRPK1 in complex with MSC2711186
To Be Published
|
|
1OVW
 
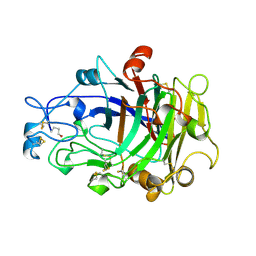 | | ENDOGLUCANASE I COMPLEXED WITH NON-HYDROLYSABLE SUBSTRATE ANALOGUE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-thio-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-1,4-dithio-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Sulzenbacher, G, Davies, G.J, Schulein, M. | | Deposit date: | 1996-10-17 | | Release date: | 1997-10-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Fusarium oxysporum endoglucanase I with a nonhydrolyzable substrate analogue: substrate distortion gives rise to the preferred axial orientation for the leaving group.
Biochemistry, 35, 1996
|
|
1OF9
 
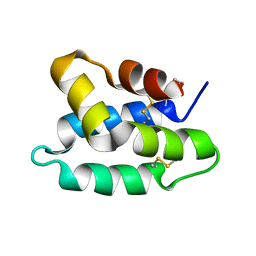 | | Solution structure of the pore forming toxin of entamoeba histolytica (Amoebapore A) | | Descriptor: | PORE-FORMING PEPTIDE AMEOBAPORE A | | Authors: | Hecht, O, Schleinkofer, K, Bruhn, H, Leippe, M, Van Nuland, N, Dingley, A.J, Grotzinger, J. | | Deposit date: | 2003-04-09 | | Release date: | 2004-02-26 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Pore-Forming Protein of Entamoeba Histolytica
J.Biol.Chem., 279, 2004
|
|
3A3H
 
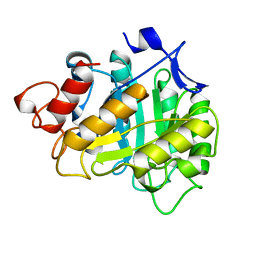 | | CELLOTRIOSE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHERANS AT 1.6 A RESOLUTION | | Descriptor: | ENDOGLUCANASE, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Davies, G.J, Brzozowski, A.M, Andersen, K, Schulein, M. | | Deposit date: | 1998-02-01 | | Release date: | 1999-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase.
Biochemistry, 37, 1998
|
|
2LRD
 
 | | The solution structure of the monomeric Acanthaporin | | Descriptor: | Acanthaporin | | Authors: | Michalek, M, Soennichsen, F.D, Wechselberger, R, Dingley, A.J, Wienk, H, Simanski, M, Herbst, R, Lorenzen, I, Marciano-Cabral, F, Gelhaus, C, Groetzinger, J, Leippe, M. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and function of a unique pore-forming protein from a pathogenic acanthamoeba.
Nat.Chem.Biol., 9, 2013
|
|
1DYM
 
 | | Humicola insolens Endocellulase Cel7B (EG 1) E197A Mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Davies, G.J, Moraz, O, Driguez, H, Schulein, M. | | Deposit date: | 2000-02-03 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the family 7 endoglucanase I (Cel7B) from Humicola insolens at 2.2 A resolution and identification of the catalytic nucleophile by trapping of the covalent glycosyl-enzyme intermediate.
Biochem.J., 335 ( Pt 2), 1998
|
|
5AJ3
 
 | | Structure of the small subunit of the mammalian mitoribosome | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MITORIBOSOMAL 12S RRNA, ... | | Authors: | Greber, B.J, Bieri, P, Leibundgut, M, Leitner, A, Aebersold, R, Boehringer, D, Ban, N. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-22 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome.
Science, 348, 2015
|
|
5AJ4
 
 | | Structure of the 55S mammalian mitoribosome. | | Descriptor: | 28S RIBOSOMAL PROTEIN S18B, MITOCHONDRIAL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Greber, B.J, Bieri, P, Leibundgut, M, Leitner, A, Aebersold, R, Boehringer, D, Ban, N. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-22 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The complete structure of the 55S mammalian mitochondrial ribosome.
Science, 348, 2015
|
|
4CE8
 
 | | Perdeuterated Pseudomonas aeruginosa Lectin II complex with hydrogenated L-Fucose and Calcium | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, SULFATE ION, ... | | Authors: | Cuypers, M.G, Mitchell, E.P, Mossou, E, Pokorna, M, Wimmerova, M, Imberty, A, Moulin, M, Haertlein, M, Forsyth, V.T. | | Deposit date: | 2013-11-10 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Perdeuterated Pseudomonas Aeruginosa Lectin II Complex with Hydrogenated L Fucose and Calcium
To be Published
|
|
1QH6
 
 | | CATALYSIS AND SPECIFICITY IN ENZYMATIC GLYCOSIDE HYDROLASES: A 2,5B CONFORMATION FOR THE GLYCOSYL-ENZYME INTERMIDIATE REVEALED BY THE STRUCTURE OF THE BACILLUS AGARADHAERENS FAMILY 11 XYLANASE | | Descriptor: | XYLANASE, beta-D-xylopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-xylopyranose | | Authors: | Sabini, E, Sulzenbacher, G, Dauter, M, Dauter, Z, Jorgensen, P.L, Schulein, M, Dupont, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 1999-05-11 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalysis and specificity in enzymatic glycoside hydrolysis: a 2,5B conformation for the glycosyl-enzyme intermediate revealed by the structure of the Bacillus agaradhaerens family 11 xylanase.
Chem.Biol., 6, 1999
|
|
1HCE
 
 | | STRUCTURE OF HISACTOPHILIN IS SIMILAR TO INTERLEUKIN-1 BETA AND FIBROBLAST GROWTH FACTOR | | Descriptor: | HISACTOPHILIN | | Authors: | Habazettl, J, Gondol, D, Wiltscheck, R, Otlewski, J, Schleicher, M, Holak, T.A. | | Deposit date: | 1994-07-12 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of hisactophilin is similar to interleukin-1 beta and fibroblast growth factor.
Nature, 359, 1992
|
|
2OWI
 
 | | Solution structure of the RGS domain from human RGS18 | | Descriptor: | Regulator of G-protein signaling 18 | | Authors: | Higman, V.A, Leidert, M, Bray, J, Elkins, J, Soundararajan, M, Doyle, D.A, Gileadi, C, Phillips, C, Schoch, G, Yang, X, Brockmann, C, Schmieder, P, Diehl, A, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Oschkinat, H, Ball, L.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-16 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1QH7
 
 | | CATALYSIS AND SPECIFICITY IN ENZYMATIC GLYCOSIDE HYDROLASES: A 2,5B CONFORMATION FOR THE GLYCOSYL-ENZYME INTERMIDIATE REVEALED BY THE STRUCTURE OF THE BACILLUS AGARADHAERENS FAMILY 11 XYLANASE | | Descriptor: | XYLANASE, beta-D-xylopyranose | | Authors: | Sabini, E, Sulzenbacher, G, Dauter, M, Dauter, Z, Jorgensen, P.L, Schulein, M, Dupont, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 1999-05-11 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Catalysis and specificity in enzymatic glycoside hydrolysis: a 2,5B conformation for the glycosyl-enzyme intermediate revealed by the structure of the Bacillus agaradhaerens family 11 xylanase.
Chem.Biol., 6, 1999
|
|
1HCD
 
 | | STRUCTURE OF HISACTOPHILIN IS SIMILAR TO INTERLEUKIN-1 BETA AND FIBROBLAST GROWTH FACTOR | | Descriptor: | HISACTOPHILIN | | Authors: | Habazettl, J, Gondol, D, Wiltscheck, R, Otlewski, J, Schleicher, M, Holak, T.A. | | Deposit date: | 1994-05-03 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of hisactophilin is similar to interleukin-1 beta and fibroblast growth factor.
Nature, 359, 1992
|
|
5LFQ
 
 | | Crystal Structure of the Bacterial Proteasome Activator Bpa of Mycobacterium tuberculosis (space group P3) | | Descriptor: | Bacterial proteasome activator | | Authors: | Bolten, M, Delley, C.L, Leibundgut, M, Boehringer, D, Ban, N, Weber-Ban, E. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Structural Analysis of the Bacterial Proteasome Activator Bpa in Complex with the 20S Proteasome.
Structure, 24, 2016
|
|
1DYS
 
 | | Endoglucanase CEL6B from Humicola insolens | | Descriptor: | ENDOGLUCANASE | | Authors: | Davies, G.J, Brzozowski, A.M, Dauter, M, Varrot, A, Schulein, M. | | Deposit date: | 2000-02-08 | | Release date: | 2001-02-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Function of Humicola Insolens Family 6 Cellulases: Structure of the Endoglucanase, Cel6B, at 1.6 A Resolution
Biochem.J., 348, 2000
|
|
3K2S
 
 | | Solution structure of double super helix model | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Apolipoprotein A-I, CHOLESTEROL | | Authors: | Wu, Z, Gogonea, V, Lee, X, Wagner, M.A, Li, X.-M, Huang, Y, Undurti, A, May, R.P, Haertlein, M, Moulin, M, Gutsche, I, Zaccai, G, Didonato, J.A, Hazen, L.S. | | Deposit date: | 2009-09-30 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Double superhelix model of high density lipoprotein.
J.Biol.Chem., 284, 2009
|
|
5CM1
 
 | | X-ray structure of perdeuterated TTR mutant - T119M at 1.22A | | Descriptor: | Transthyretin | | Authors: | Yee, A.W, Moulin, M, Mossou, E, Haertlein, M, Mitchell, E.P, Cooper, J.B, Forsyth, V.T. | | Deposit date: | 2015-07-16 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | X-ray structure of perdeuterated TTR mutant - T119M at 1.22A
To Be Published
|
|
5LFJ
 
 | | Crystal Structure of the Bacterial Proteasome Activator Bpa of Mycobacterium tuberculosis | | Descriptor: | Bacterial proteasome activator | | Authors: | Bolten, M, Delley, C.L, Leibundgut, M, Boehringer, D, Ban, N, Weber-Ban, E. | | Deposit date: | 2016-07-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis of the Bacterial Proteasome Activator Bpa in Complex with the 20S Proteasome.
Structure, 24, 2016
|
|
5LFP
 
 | | Crystal Structure of the Bacterial Proteasome Activator Bpa of Mycobacterium tuberculosis (space group P6322, SeMet) | | Descriptor: | Bacterial proteasome activator | | Authors: | Bolten, M, Delley, C.L, Leibundgut, M, Boehringer, D, Ban, N, Weber-Ban, E. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-23 | | Last modified: | 2016-12-14 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | Structural Analysis of the Bacterial Proteasome Activator Bpa in Complex with the 20S Proteasome.
Structure, 24, 2016
|
|
5LZP
 
 | | Binding of the C-terminal GQYL motif of the bacterial proteasome activator Bpa to the 20S proteasome | | Descriptor: | Bacterial proteasome activator, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Bolten, M, Delley, C.L, Leibundgut, M, Boehringer, D, Ban, N, Weber-Ban, E. | | Deposit date: | 2016-09-30 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Analysis of the Bacterial Proteasome Activator Bpa in Complex with the 20S Proteasome.
Structure, 24, 2016
|
|
5LRT
 
 | | Structure of the Deamidase-Depupylase Dop of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ADP and Phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, Depupylase, ... | | Authors: | Bolten, M, Vahlensieck, C, Lipp, C, Leibundgut, M, Ban, N, Weber-Ban, E. | | Deposit date: | 2016-08-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Depupylase Dop Requires Inorganic Phosphate in the Active Site for Catalysis.
J. Biol. Chem., 292, 2017
|
|
5CLX
 
 | | X-ray structure of perdeuterated TTR mutant - S52P at 1.28A resolution | | Descriptor: | GLYCEROL, Transthyretin | | Authors: | Yee, A.W, Moulin, M, Mossou, E, Haertlein, M, Mitchell, E.P, Cooper, J.B, Forsyth, V.T. | | Deposit date: | 2015-07-16 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | X-ray structure of perdeuterated TTR mutant - S52P at 1.28A resolution
To Be Published
|
|
5MMM
 
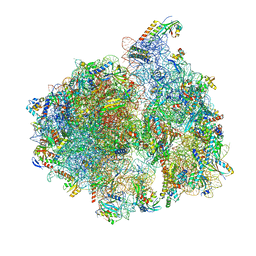 | | Structure of the 70S chloroplast ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein 2, ... | | Authors: | Bieri, P, Leibundgut, M, Saurer, M, Boehringer, D, Ban, N. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The complete structure of the chloroplast 70S ribosome in complex with translation factor pY.
EMBO J., 36, 2017
|
|
5CLZ
 
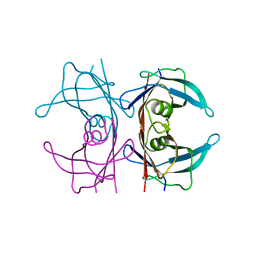 | | X-ray structure of TTR mutant - T119M at 1.22A resolution | | Descriptor: | Transthyretin | | Authors: | Yee, A.W, Moulin, M, Mossou, E, Cooper, J.B, Haertlein, M, Mitchell, E.P, Forsyth, V.T. | | Deposit date: | 2015-07-16 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | X-ray structure of TTR mutant - T119M at 1.22A resolution
To Be Published
|
|
