7TB7
 
 | | Crystal structure of D179N KPC-2 beta-lactamase | | Descriptor: | CITRIC ACID, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | van den Akker, F, Alsenani, T. | | Deposit date: | 2021-12-21 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Structural Characterization of the D179N and D179Y Variants of KPC-2 beta-Lactamase: Omega-Loop Destabilization as a Mechanism of Resistance to Ceftazidime-Avibactam.
Antimicrob.Agents Chemother., 66, 2022
|
|
7TBX
 
 | | Crystal structure of D179Y KPC-2 beta-lactamase | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | van den Akker, F, Alsenani, T. | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Structural Characterization of the D179N and D179Y Variants of KPC-2 beta-Lactamase: Omega-Loop Destabilization as a Mechanism of Resistance to Ceftazidime-Avibactam.
Antimicrob.Agents Chemother., 66, 2022
|
|
7TC1
 
 | |
6SWQ
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH iBET-BD2 (GSK046) | | Descriptor: | 1,2-ETHANEDIOL, 4-acetamido-3-fluoranyl-~{N}-(4-oxidanylcyclohexyl)-5-[(1~{S})-1-phenylethoxy]benzamide, Bromodomain-containing protein 4 | | Authors: | Chung, C. | | Deposit date: | 2019-09-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation.
Science, 368, 2020
|
|
6SWP
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH iBET-BD2 (GSK046) | | Descriptor: | 1,2-ETHANEDIOL, 4-acetamido-3-fluoranyl-~{N}-(4-oxidanylcyclohexyl)-5-[(1~{S})-1-phenylethoxy]benzamide, Bromodomain-containing protein 2 | | Authors: | Chung, C. | | Deposit date: | 2019-09-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation.
Science, 368, 2020
|
|
7USG
 
 | | BRD2-BD2 in complex with MDP5 | | Descriptor: | (8M)-8-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-(morpholin-4-yl)-4H-1-benzopyran-4-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 2, ... | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Targeting BRD4 and PI3K signaling pathways for the treatment of medulloblastoma.
J Control Release, 354, 2023
|
|
7USI
 
 | | BRD2-BD1 in complex with MDP5 | | Descriptor: | (8M)-8-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-(morpholin-4-yl)-4H-1-benzopyran-4-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 2, ... | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting BRD4 and PI3K signaling pathways for the treatment of medulloblastoma.
J Control Release, 354, 2023
|
|
7USH
 
 | | BRD2-BD2 in complex with SF2523 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-(morpholin-4-yl)-7H-thieno[3,2-b]pyran-7-one, Bromodomain-containing protein 2 | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Targeting BRD4 and PI3K signaling pathways for the treatment of medulloblastoma.
J Control Release, 354, 2023
|
|
6SWO
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH iBET-BD1 (GSK778) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(methoxymethyl)-1-[(1~{R})-1-phenylethyl]-8-[[(3~{S})-pyrrolidin-3-yl]methoxy]imidazo[4,5-c]quinolin-7-yl]-3,5-dimethyl-1,2-oxazole, Bromodomain-containing protein 2 | | Authors: | Chung, C. | | Deposit date: | 2019-09-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation.
Science, 368, 2020
|
|
7USJ
 
 | | BRD4-BD2 in complex with SF2523 | | Descriptor: | 3-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-(morpholin-4-yl)-7H-thieno[3,2-b]pyran-7-one, Bromodomain-containing protein 4 | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Targeting BRD4 and PI3K signaling pathways for the treatment of medulloblastoma.
J Control Release, 354, 2023
|
|
7USK
 
 | | BRD4-BD2 Ligand free | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Targeting BRD4 and PI3K signaling pathways for the treatment of medulloblastoma.
J Control Release, 354, 2023
|
|
7LLB
 
 | | Crystal structure of KPC-2 S70G/T215P mutant with hydrolyzed meropenem | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Furey, I, Palzkill, T, Sankaran, B, Hu, L, Prasad, B.V.V. | | Deposit date: | 2021-02-03 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Local interactions with the Glu166 base and the conformation of an active site loop play key roles in carbapenem hydrolysis by the KPC-2 beta-lactamase.
J.Biol.Chem., 296, 2021
|
|
7LJK
 
 | | Crystal structure of the deacylation deficient KPC-2 F72Y mutant | | Descriptor: | Beta-lactamase | | Authors: | Furey, I, Palzkill, T, Sankaran, B, Hu, L, Prasad, B.V.V. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Local interactions with the Glu166 base and the conformation of an active site loop play key roles in carbapenem hydrolysis by the KPC-2 beta-lactamase.
J.Biol.Chem., 296, 2021
|
|
7LLH
 
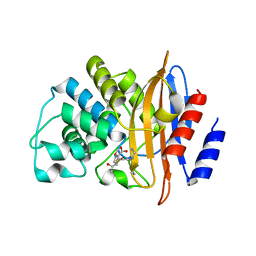 | | KPC-2 F72Y mutant with acylated imipenem | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Furey, I, Palzkill, T, Sankaran, B, Hu, L, Prasad, B.V.V. | | Deposit date: | 2021-02-03 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Local interactions with the Glu166 base and the conformation of an active site loop play key roles in carbapenem hydrolysis by the KPC-2 beta-lactamase.
J.Biol.Chem., 296, 2021
|
|
6SWN
 
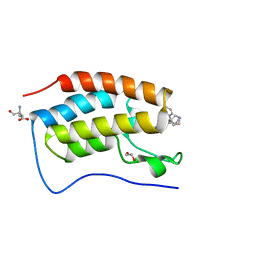 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH iBET-BD1 (GSK778) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(methoxymethyl)-1-[(1~{R})-1-phenylethyl]-8-[[(3~{S})-pyrrolidin-3-yl]methoxy]imidazo[4,5-c]quinolin-7-yl]-3,5-dimethyl-1,2-oxazole, Bromodomain-containing protein 4, ... | | Authors: | Chung, C. | | Deposit date: | 2019-09-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation.
Science, 368, 2020
|
|
2PTV
 
 | | Structure of NK cell receptor ligand CD48 | | Descriptor: | CD48 antigen | | Authors: | Deng, L, Velikovsky, C.A, Mariuzza, R.A. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of natural killer receptor 2B4 bound to CD48 reveals basis for heterophilic recognition in signaling lymphocyte activation molecule family.
Immunity, 27, 2007
|
|
1AZM
 
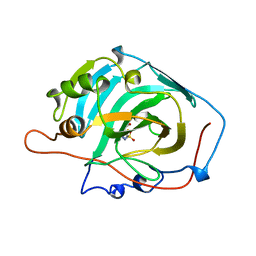 | |
3S2V
 
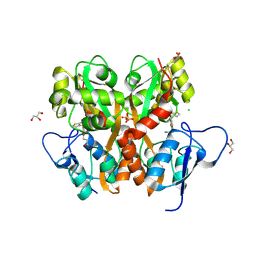 | | Crystal Structure of the Ligand Binding Domain of GluK1 in Complex with an Antagonist (S)-1-(2'-Amino-2'-carboxyethyl)-3-[(2-carboxythien-3-yl)methyl]thieno[3,4-d]pyrimidin-2,4-dione at 2.5 A Resolution | | Descriptor: | (S)-1-(2'-AMINO-2'-CARBOXYETHYL)-3-[(2-CARBOXYTHIEN-3-YL)METHYL]THIENO[3,4-D]PYRIMIDIN-2,4-DIONE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2011-05-17 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Selective kainate receptor (GluK1) ligands structurally based upon 1H-cyclopentapyrimidin-2,4(1H,3H)-dione: synthesis, molecular modeling, and pharmacological and biostructural characterization.
J.Med.Chem., 54, 2011
|
|
5VCL
 
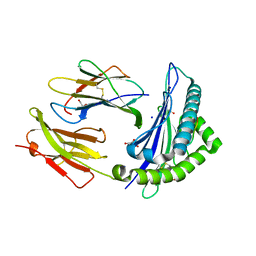 | | Structure of the Qdm peptide bound to Qa-1a | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H2-T23 protein, ... | | Authors: | Ying, G, Zajonc, D.M. | | Deposit date: | 2017-03-31 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Qa-1a with bound Qa-1 determinant modifier peptide.
PLoS ONE, 12, 2017
|
|
3V7T
 
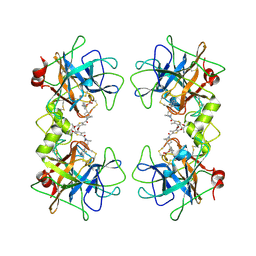 | |
6JXU
 
 | | SUMO1 bound to SLS4-SIM peptide from ICP0 | | Descriptor: | Small ubiquitin-related modifier, viral protein | | Authors: | Hembram, D.S.S, Negi, H, Shet, D, Das, R. | | Deposit date: | 2019-04-25 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Viral SUMO-Targeted Ubiquitin Ligase ICP0 is Phosphorylated and Activated by Host Kinase Chk2.
J.Mol.Biol., 432, 2020
|
|
6JXV
 
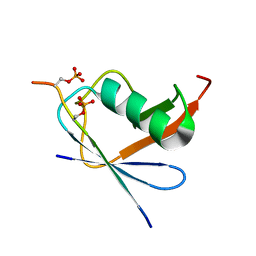 | | SUMO1 bound to phosphorylated SLS4-SIM peptide from ICP0 | | Descriptor: | Phosphorylated SLS4-SIM from ubiquitin E3 ligase ICP0, Small ubiquitin-related modifier | | Authors: | Hembram, D.S.S, Negi, H, Shet, D, Das, R. | | Deposit date: | 2019-04-25 | | Release date: | 2020-02-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Viral SUMO-Targeted Ubiquitin Ligase ICP0 is Phosphorylated and Activated by Host Kinase Chk2.
J.Mol.Biol., 432, 2020
|
|
6JXW
 
 | | Complex of SUMO2 bound SLS4 from ICP0. | | Descriptor: | SLS4-SIM from Ubiquitin E3 ligase ICP0, Small ubiquitin-related modifier 2 | | Authors: | Hembram, D.S.S, Negi, H, Shet, D, Das, R. | | Deposit date: | 2019-04-25 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Viral SUMO-Targeted Ubiquitin Ligase ICP0 is Phosphorylated and Activated by Host Kinase Chk2.
J.Mol.Biol., 432, 2020
|
|
6JXX
 
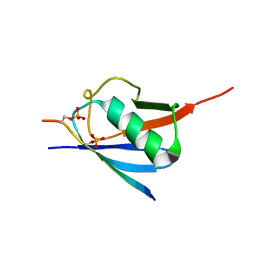 | | SUMO2 bound to phosphorylated SLS4-SIM peptide from ICP0 | | Descriptor: | Phosphorylated SLS4 from E3 ubiquitin ligase ICP0, Small ubiquitin-related modifier 2 | | Authors: | Hembram, D.S.S, Negi, H, Shet, D, Das, R. | | Deposit date: | 2019-04-25 | | Release date: | 2020-02-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Viral SUMO-Targeted Ubiquitin Ligase ICP0 is Phosphorylated and Activated by Host Kinase Chk2.
J.Mol.Biol., 432, 2020
|
|
3B7M
 
 | |
