6UQM
 
 | |
6UOJ
 
 | |
6UPM
 
 | |
6UP9
 
 | |
6UPO
 
 | |
6UQ8
 
 | |
6VWY
 
 | |
6VUE
 
 | | wild-type choline TMA lyase in complex with 1-methyl-1,2,3,6-tetrahydropyridin-3-ol | | Descriptor: | (3S)-1-methyl-1,2,3,6-tetrahydropyridin-3-ol, Choline trimethylamine-lyase, SODIUM ION | | Authors: | Ortega, M.A, Drennan, C.L. | | Deposit date: | 2020-02-15 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of a Cyclic Choline Analog That Inhibits Anaerobic Choline Metabolism by Human Gut Bacteria.
Acs Med.Chem.Lett., 11, 2020
|
|
6VWZ
 
 | |
6VXE
 
 | |
6VXC
 
 | |
6VX1
 
 | |
6VX0
 
 | |
6X5K
 
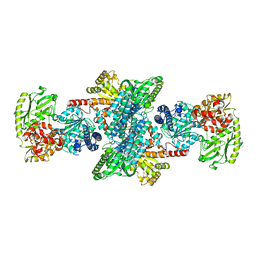 | | Crystal structure of CODH/ACS with carbon monoxide bound to the A-cluster | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, CARBON MONOXIDE, Carbon monoxide dehydrogenase/acetyl-CoA synthase subunit alpha, ... | | Authors: | Cohen, S.E, Wittenborn, E.C, Hendrickson, R, Drennan, C.L. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystallographic Characterization of the Carbonylated A-Cluster in Carbon Monoxide Dehydrogenase/Acetyl-CoA Synthase
Acs Catalysis, 10, 2020
|
|
6WTE
 
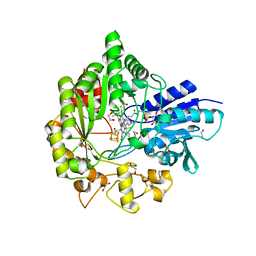 | | Structure of radical S-adenosylmethionine methyltransferase, TsrM, from Kitasatospora setae with cobalamin and [4Fe-4S] cluster bound | | Descriptor: | 1,2-ETHANEDIOL, B12-binding domain-containing protein, COBALAMIN, ... | | Authors: | Knox, H.L, Chen, P.Y.-T, Drennan, C.L, Booker, S.J. | | Deposit date: | 2020-05-02 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural basis for non-radical catalysis by TsrM, a radical SAM methylase.
Nat.Chem.Biol., 17, 2021
|
|
6WTF
 
 | | Structure of radical S-adenosylmethionine methyltransferase, TsrM, from Kitasatospora setae with tryptophan substrate and SAM analog (aza-SAM) bound | | Descriptor: | COBALAMIN, IRON/SULFUR CLUSTER, S-5'-AZAMETHIONINE-5'-DEOXYADENOSINE, ... | | Authors: | Knox, H.L, Chen, P.Y.-T, Drennan, C.L, Booker, S.J. | | Deposit date: | 2020-05-02 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis for non-radical catalysis by TsrM, a radical SAM methylase.
Nat.Chem.Biol., 17, 2021
|
|
5C8A
 
 | |
5CI4
 
 | | Ribonucleotide reductase beta subunit | | Descriptor: | MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1, beta subunit, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-10 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biophysical Characterization of Fluorotyrosine Probes Site-Specifically Incorporated into Enzymes: E. coli Ribonucleotide Reductase As an Example.
J.Am.Chem.Soc., 138, 2016
|
|
5CJW
 
 | | Isobutyryl-CoA mutase fused with bound adenosylcobalamin, GDP, Mg (holo-IcmF/GDP), and substrate pivalyl-coenzyme A | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2015-07-15 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis for Substrate Specificity in Adenosylcobalamin-dependent Isobutyryl-CoA Mutase and Related Acyl-CoA Mutases.
J.Biol.Chem., 290, 2015
|
|
5CNV
 
 | | Crystal structure of the dATP inhibited E. coli class Ia ribonucleotide reductase complex bound to GDP and TTP at 3.20 Angstroms resolution | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chen, P.Y.-T, Zimanyi, C.M, Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-18 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular basis for allosteric specificity regulation in class Ia ribonucleotide reductase from Escherichia coli.
Elife, 5, 2016
|
|
5C8E
 
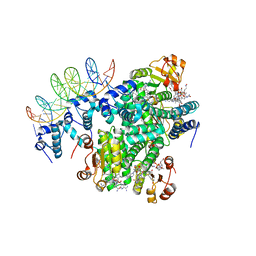 | |
5BWD
 
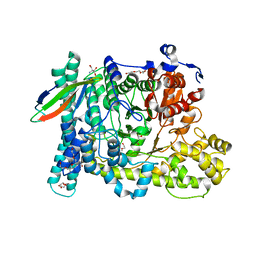 | | Benzylsuccinate alpha-gamma bound to fumarate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FUMARIC ACID, TETRAETHYLENE GLYCOL, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-06-07 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate-bound Structures of Benzylsuccinate Synthase Reveal How Toluene Is Activated in Anaerobic Hydrocarbon Degradation.
J.Biol.Chem., 290, 2015
|
|
5C4I
 
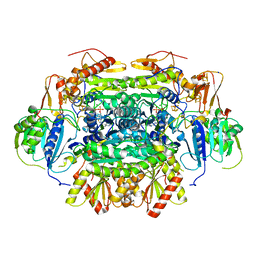 | | Structure of an Oxalate Oxidoreductase | | Descriptor: | IRON/SULFUR CLUSTER, MAGNESIUM ION, Oxalate oxidoreductase subunit alpha, ... | | Authors: | Gibson, M.I, Brignole, E.J, Pierce, E, Can, M, Ragsdale, S.W, Drennan, C.L. | | Deposit date: | 2015-06-18 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.274 Å) | | Cite: | The Structure of an Oxalate Oxidoreductase Provides Insight into Microbial 2-Oxoacid Metabolism.
Biochemistry, 54, 2015
|
|
5C8D
 
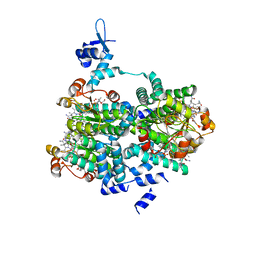 | |
5CI3
 
 | | Ribonucleotide reductase Y122 2,3,5-F3Y variant | | Descriptor: | MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1 subunit beta, SULFATE ION | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-10 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Biophysical Characterization of Fluorotyrosine Probes Site-Specifically Incorporated into Enzymes: E. coli Ribonucleotide Reductase As an Example.
J.Am.Chem.Soc., 138, 2016
|
|
