8XFB
 
 | |
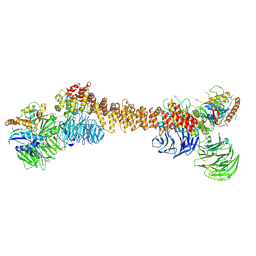
8Z9M
 
 | |
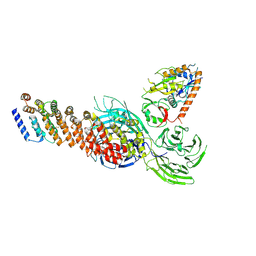
2NZ0
 
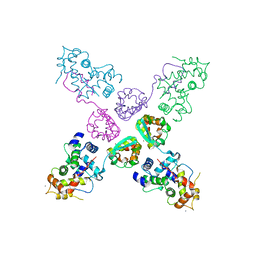 | | Crystal structure of potassium channel Kv4.3 in complex with its regulatory subunit KChIP1 | | Descriptor: | CALCIUM ION, Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 3, ... | | Authors: | Wang, H, Yan, Y, Shen, Y, Chen, L, Wang, K. | | Deposit date: | 2006-11-22 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for modulation of Kv4 K(+) channels by auxiliary KChIP subunits.
Nat.Neurosci., 10, 2007
|
|
9AUJ
 
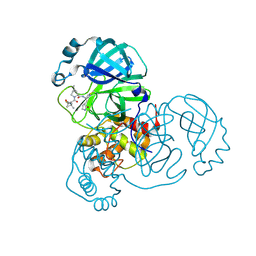 | | Structure of SARS-CoV-2 Mpro mutant (S144A) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.486 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUM
 
 | | Structure of SARS-CoV-2 Mpro mutant (T21I,S144A,T304I) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUK
 
 | | Structure of SARS-CoV-2 Mpro mutant (A173V) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUO
 
 | | Structure of SARS-CoV-2 Mpro mutant (L50F,T304I) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUN
 
 | | Structure of SARS-CoV-2 Mpro mutant (T21I,T304I) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUL
 
 | | Structure of SARS-CoV-2 Mpro mutant (A173V,T304I)) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.421 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
7KFA
 
 | | PCSK9 in complex with PCSK9i a 13mer cyclic peptide LDLR disruptor | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Chopra, R, Xu, M, Spraggon, G. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification of a PCSK9-LDLR disruptor peptide with in vivo function.
Cell Chem Biol, 29, 2022
|
|
7KEV
 
 | | PCSK9 in complex with a cyclic peptide LDLR disruptor | | Descriptor: | CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, Proprotein convertase subtilisin/kexin type 9 Propeptide, ... | | Authors: | Spraggon, G, Chopra, R. | | Deposit date: | 2020-10-12 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a PCSK9-LDLR disruptor peptide with in vivo function.
Cell Chem Biol, 29, 2022
|
|
5GUF
 
 | |
7RFR
 
 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-(4-methoxy-1H-indole-2-carbonyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Gajiwala, K.S, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.626 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
7RFU
 
 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-[N-(methanesulfonyl)-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2022-01-05 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
7RFW
 
 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2022-01-05 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
7RFS
 
 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2022-01-05 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
7RLZ
 
 | |
7RM1
 
 | |
7RLX
 
 | |
7RLV
 
 | |
7RLW
 
 | |
7RM0
 
 | |
7RLY
 
 | |
7RM3
 
 | | Antibody 2E10.E9 in complex with P. vivax CSP peptide ANGAGNQPGANGAGNQPGANGAGGQAA | | Descriptor: | 2E10.E9 Fab heavy chain, 2E10.E9 Fab light chain, ACETATE ION, ... | | Authors: | Kucharska, I, Ivanochko, D, Julien, J.P. | | Deposit date: | 2021-07-26 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis of Plasmodium vivax inhibition by antibodies binding to the circumsporozoite protein repeats.
Elife, 11, 2022
|
|
7YNX
 
 | | Crystal structure of Pirh2 bound to poly-Ala peptide | | Descriptor: | RING finger and CHY zinc finger domain-containing protein 1, SODIUM ION, SULFATE ION, ... | | Authors: | Dong, C, Yan, X, Li, Y. | | Deposit date: | 2022-08-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of an Ala-rich C-degron by the E3 ligase Pirh2.
Nat Commun, 14, 2023
|
|
