7MBU
 
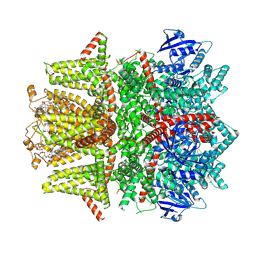 | | Cryo-EM structure of zebrafish TRPM5 E337A mutant in the presence of 5 mM calcium (high calcium occupancy in the transmembrane domain) | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MBV
 
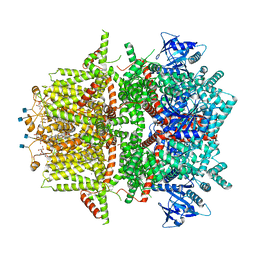 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 5 mM calcium and 0.5 mM NDNA | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MBP
 
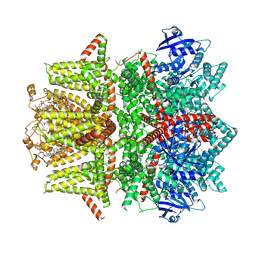 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 1 mM EDTA | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6K05
 
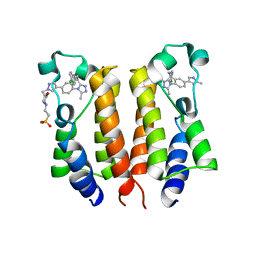 | | Crystal structure of BRD2(BD1)with ligand BY27 bound | | Descriptor: | (6~{R})-~{N}-(4-chlorophenyl)-1-methyl-8-(1-methylpyrazol-4-yl)-5,6-dihydro-4~{H}-[1,2,4]triazolo[4,3-a][1]benzazepin-6-amine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2 | | Authors: | Lu, T, Lu, W, Chen, D, Zhao, Y, Luo, C. | | Deposit date: | 2019-05-05 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Discovery, structural insight, and bioactivities of BY27 as a selective inhibitor of the second bromodomains of BET proteins.
Eur.J.Med.Chem., 182, 2019
|
|
6K04
 
 | | Crystal structure of BRD2(BD2)with ligand BY27 bound | | Descriptor: | (6~{R})-~{N}-(4-chlorophenyl)-1-methyl-8-(1-methylpyrazol-4-yl)-5,6-dihydro-4~{H}-[1,2,4]triazolo[4,3-a][1]benzazepin-6-amine, Bromodomain-containing protein 2 | | Authors: | Lu, T, Lu, W, Chen, D, Zhao, Y, Luo, C. | | Deposit date: | 2019-05-05 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Discovery, structural insight, and bioactivities of BY27 as a selective inhibitor of the second bromodomains of BET proteins.
Eur.J.Med.Chem., 182, 2019
|
|
4M0I
 
 | |
4PBV
 
 | | Crystal structure of chicken receptor protein tyrosine phosphatase sigma in complex with TrkC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NT-3 growth factor receptor, Protein-tyrosine phosphatase CRYPalpha1 isoform, ... | | Authors: | Coles, C.H, Mitakidis, N, Zhang, P, Elegheert, J, Lu, W, Stoker, A.W, Nakagawa, T, Craig, A.M, Jones, E.Y, Aricescu, A.R. | | Deposit date: | 2014-04-14 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for extracellular cis and trans RPTP sigma signal competition in synaptogenesis.
Nat Commun, 5, 2014
|
|
4PBX
 
 | | Crystal structure of the six N-terminal domains of human receptor protein tyrosine phosphatase sigma | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase S | | Authors: | Coles, C.H, Mitakidis, N, Zhang, P, Elegheert, J, Lu, W, Stoker, A.W, Nakagawa, T, Craig, A.M, Jones, E.Y, Aricescu, A.R. | | Deposit date: | 2014-04-14 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for extracellular cis and trans RPTP sigma signal competition in synaptogenesis.
Nat Commun, 5, 2014
|
|
4PBW
 
 | | Crystal structure of chicken receptor protein tyrosine phosphatase sigma in complex with TrkC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NT-3 growth factor receptor, Protein-tyrosine phosphatase CRYPalpha1 isoform | | Authors: | Coles, C.H, Mitakidis, N, Zhang, P, Elegheert, J, Lu, W, Stoker, A.W, Nakagawa, T, Craig, A.M, Jones, E.Y, Aricescu, A.R. | | Deposit date: | 2014-04-14 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis for extracellular cis and trans RPTP sigma signal competition in synaptogenesis.
Nat Commun, 5, 2014
|
|
4R7L
 
 | | Structure of Human Leukotriene A4 Hydrolase in complex with inhibitor H1 | | Descriptor: | ACETATE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Ouyang, P, Cui, K, Lu, W, Huang, J. | | Deposit date: | 2014-08-27 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Human Leukotriene A4 Hydrolase in complex with SAHA
To be Published
|
|
7YOA
 
 | |
7N8T
 
 | | Crystal Structure of AMP-bound Human JNK2 | | Descriptor: | ADENOSINE MONOPHOSPHATE, HEXAETHYLENE GLYCOL, Mitogen-activated protein kinase 9 | | Authors: | Li, L, Gurbani, D, Westover, K.D. | | Deposit date: | 2021-06-15 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Development of a Covalent Inhibitor of c-Jun N-Terminal Protein Kinase (JNK) 2/3 with Selectivity over JNK1.
J.Med.Chem., 66, 2023
|
|
4FC4
 
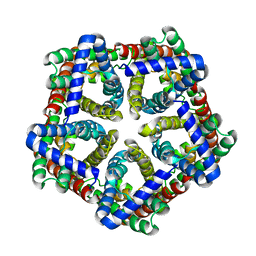 | | FNT family ion channel | | Descriptor: | Nitrite transporter NirC, octyl beta-D-glucopyranoside | | Authors: | Lue, W, Schwarzer, N, Du, J, Gerbig-Smentek, E, Andrade, S.L.A, Einsle, O. | | Deposit date: | 2012-05-24 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of the nitrite channel NirC from Salmonella typhimurium.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8ELC
 
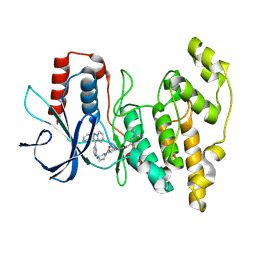 | | Human JNK2 bound to covalent inhibitor YL2056 | | Descriptor: | 4-(dimethylamino)-N-{4-[(3S)-3-({4-[(8R)-2-phenylpyrazolo[1,5-a]pyridin-3-yl]pyrimidin-2-yl}amino)pyrrolidine-1-carbonyl]phenyl}butanamide, Mitogen-activated protein kinase 9 | | Authors: | Li, L, Gurbani, D, Westover, K.D. | | Deposit date: | 2022-09-23 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.072 Å) | | Cite: | Development of a Covalent Inhibitor of c-Jun N-Terminal Protein Kinase (JNK) 2/3 with Selectivity over JNK1.
J.Med.Chem., 66, 2023
|
|
3Q7K
 
 | | Formate Channel FocA from Salmonella typhimurium | | Descriptor: | FORMIC ACID, Probable formate transporter | | Authors: | Lue, W, Du, J, Wacker, T, Gerbig-Smentek, E, Andrade, S.L.A, Einsle, O. | | Deposit date: | 2011-01-05 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | pH-dependent gating in a FocA formate channel
Science, 332, 2011
|
|
2ZJJ
 
 | |
2ZJM
 
 | |
2ZJI
 
 | |
2ZJN
 
 | |
2ZJH
 
 | |
3GNY
 
 | | Crystal structure of human alpha-defensin 1 (HNP1) | | Descriptor: | CHLORIDE ION, GLYCEROL, Neutrophil defensin 1 | | Authors: | Pazgier, M, Lu, W.-Y. | | Deposit date: | 2009-03-18 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Through the looking glass, mechanistic insights from enantiomeric human defensins.
J.Biol.Chem., 284, 2009
|
|
3TPX
 
 | |
3GO0
 
 | |
4RXZ
 
 | |
6WYS
 
 | | Lon protease proteolytic domain | | Descriptor: | Lon protease homolog, mitochondrial, SULFATE ION | | Authors: | Lee, C.C, Spraggon, G. | | Deposit date: | 2020-05-13 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.229 Å) | | Cite: | Structure-Based Design of Selective LONP1 Inhibitors for Probing In Vitro Biology.
J.Med.Chem., 64, 2021
|
|
