5TKE
 
 | | Crystal Structure of Eukaryotic Hydrolase | | Descriptor: | O-GlcNAcase: chimera construct | | Authors: | Li, B, Jiang, J. | | Deposit date: | 2016-10-06 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | Structures of human O-GlcNAcase and its complexes reveal a new substrate recognition mode.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6IGB
 
 | | the structure of Pseudomonas aeruginosa Periplasmic gluconolactonase, PpgL | | Descriptor: | ACETATE ION, Periplasmic gluconolactonase, PpgL, ... | | Authors: | Song, Y.J, Shen, Y.L, Wang, K.L, Li, T, Zhu, Y.B, Li, C.C, He, L.H, Zhao, N.L, Zhao, C, Yang, J, Huang, Q, Mu, X.Y. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural and Functional Insights into PpgL, a Metal-Independent beta-Propeller Gluconolactonase That Contributes toPseudomonas aeruginosaVirulence.
Infect.Immun., 87, 2019
|
|
5UN8
 
 | |
7UDS
 
 | | Structure of lineage I (Pinneo) Lassa virus glycoprotein bound to Fab 25.10C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 25.10C Fab Heavy Chain, ... | | Authors: | Buck, T.K, Enriquez, A.E, Hastie, K.M. | | Deposit date: | 2022-03-20 | | Release date: | 2022-06-15 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Neutralizing Antibodies against Lassa Virus Lineage I.
Mbio, 13, 2022
|
|
7UL7
 
 | | Lineage I (Pinneo) Lassa virus glycoprotein bound to 18.5C-M30 Fab | | Descriptor: | 18.5C-M30 Fab Heavy Chain, 18.5C-M30 Fab Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Buck, T.K, Enriquez, A.S, Hastie, K.M. | | Deposit date: | 2022-04-04 | | Release date: | 2022-06-15 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Neutralizing Antibodies against Lassa Virus Lineage I.
Mbio, 13, 2022
|
|
5UN9
 
 | | The crystal structure of human O-GlcNAcase in complex with Thiamet-G | | Descriptor: | (3AR,5R,6S,7R,7AR)-2-(ETHYLAMINO)-5-(HYDROXYMETHYL)-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]THIAZOLE-6,7-DIOL, Protein O-GlcNAcase | | Authors: | Li, B, Jiang, J. | | Deposit date: | 2017-01-30 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of human O-GlcNAcase and its complexes reveal a new substrate recognition mode.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6J0Z
 
 | | Crystal structure of AlpK | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative angucycline-like polyketide oxygenase | | Authors: | Wang, W, Liu, Y, Liang, H. | | Deposit date: | 2018-12-27 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | Crystal structure of AlpK: An essential monooxygenase involved in the biosynthesis of kinamycin
Biochem. Biophys. Res. Commun., 510, 2019
|
|
8T05
 
 | | Structure of Ciona Myomaker bound to Fab1A1 | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1A1 Fab heavy chain, 1A1 Fab light chain, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2023-05-31 | | Release date: | 2023-09-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structures of Myomaker reveal a molecular basis for myoblast fusion.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T07
 
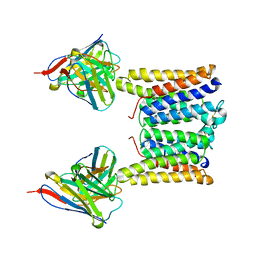 | | Structure of mouse Myomaker mutant-Y118A bound to Fab18G7 | | Descriptor: | 18G7 Fab heavy chain, 18G7 Fab light chain, Protein myomaker, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2023-05-31 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structures of Myomaker reveal a molecular basis for myoblast fusion.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T06
 
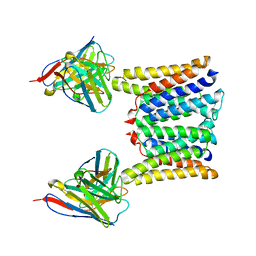 | | Structure of mouse Myomaker mutant-R107A bound to Fab18G7 | | Descriptor: | 18G7 Fab heavy chain, 18G7 Fab light chain, Protein myomaker, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2023-05-31 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM structures of Myomaker reveal a molecular basis for myoblast fusion.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T03
 
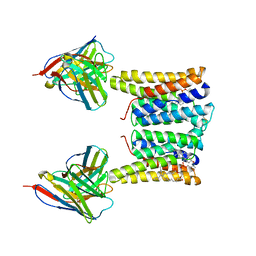 | | Structure of mouse Myomaker bound to Fab18G7 in detergent | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 18G7 Fab heavy chain, 18G7 Fab light chain, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2023-05-31 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Cryo-EM structures of Myomaker reveal a molecular basis for myoblast fusion.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T04
 
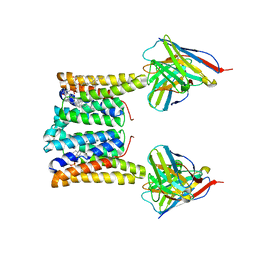 | | Structure of mouse Myomaker bound to Fab18G7 in nanodiscs | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 18G7 Fab heavy chain, 18G7 Fab light chain, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2023-05-31 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Cryo-EM structures of Myomaker reveal a molecular basis for myoblast fusion.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6OWN
 
 | |
4Y6K
 
 | | Complex structure of presenilin homologue PSH bound to an inhibitor | | Descriptor: | N-{(2R,4S,5S)-2-benzyl-5-[(tert-butoxycarbonyl)amino]-4-hydroxy-6-phenylhexanoyl}-L-leucyl-L-phenylalaninamide, Uncharacterized protein PSH | | Authors: | Dang, S, Wu, S, Wang, J, Shi, Y. | | Deposit date: | 2015-02-13 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.855 Å) | | Cite: | Cleavage of amyloid precursor protein by an archaeal presenilin homologue PSH
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1MLM
 
 | |
1MLH
 
 | |
3JSF
 
 | |
5XNV
 
 | | Crystal structure of YEATS2 YEATS bound to H3K27ac peptide | | Descriptor: | ALA-ALA-ARG-ALY-SER-ALA-PRO-ALA, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Li, H.T, Guan, H.P, Zhao, D. | | Deposit date: | 2017-05-24 | | Release date: | 2017-11-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | YEATS2 links histone acetylation to tumorigenesis of non-small cell lung cancer.
Nat Commun, 8, 2017
|
|
3JTU
 
 | |
3JSG
 
 | |
1MLN
 
 | |
5Z2N
 
 | | Structure of Orp1L N-terminal Domain | | Descriptor: | Oxysterol-binding protein-related protein 1 | | Authors: | Ma, X.L, Liu, K, Li, J, Li, H.H, Li, J, Yang, C.L, Liang, H.H. | | Deposit date: | 2018-01-03 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A non-canonical GTPase interaction enables ORP1L-Rab7-RILP complex formation and late endosome positioning.
J. Biol. Chem., 293, 2018
|
|
1MLS
 
 | |
1MLG
 
 | |
1MLR
 
 | |
