6FMR
 
 | | IMISX-EP of Se-PepTSt | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Di-or tripeptide:H+ symporter, PHOSPHATE ION | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-29 | | Last modified: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6FMW
 
 | | IMISX-EP of Hg-BacA cocrystallization | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MERCURY (II) ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6FMS
 
 | | IMISX-EP of Se-LspA | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Globomycin, Lipoprotein signal peptidase | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6FMT
 
 | | IMISX-EP of Hg-BacA Soaking SAD | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MERCURY (II) ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6FMY
 
 | | IMISX-EP of S-PepTSt | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Di-or tripeptide:H+ symporter, ... | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6FMX
 
 | | IMISX-EP of W-PgpB | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Phosphatidylglycerophosphatase B, TUNGSTATE(VI)ION | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
4ACM
 
 | | CDK2 IN COMPLEX WITH 3-AMINO-6-(4-{[2-(DIMETHYLAMINO)ETHYL]SULFAMOYL}-PHENYL)-N-PYRIDIN-3-YLPYRAZINE-2-CARBOXAMIDE | | Descriptor: | 3-AMINO-6-(4-{[2-(DIMETHYLAMINO)ETHYL]SULFAMOYL}PHENYL)-N-PYRIDIN-3-YLPYRAZINE-2-CARBOXAMIDE, CYCLIN-DEPENDENT KINASE 2, GLYCEROL | | Authors: | Berg, S, Bhat, R, Anderson, M, Bergh, M, Brassington, C, Hellberg, S, Jerning, E, Hogdin, K, Lo-Alfredsson, Y, Neelissen, J, Nilsson, Y, Ormo, M, Soderman, P, Stanway, J, Tucker, J, von Berg, S, Weigelt, T, Xue, Y. | | Deposit date: | 2011-12-16 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery of novel potent and highly selective glycogen synthase kinase-3 beta (GSK3 beta ) inhibitors for Alzheimer's disease: design, synthesis, and characterization of pyrazines.
J. Med. Chem., 55, 2012
|
|
3IXV
 
 | | Scorpion Hemocyanin resting state pseudo atomic model built based on cryo-EM density map | | Descriptor: | Hemocyanin AA6 chain | | Authors: | Cong, Y, Zhang, Q, Woolford, D, Schweikardt, T, Khant, H, Ludtke, S, Chiu, W, Decker, H. | | Deposit date: | 2009-02-13 | | Release date: | 2009-06-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structural Mechanism of SDS-Induced Enzyme Activity of Scorpion Hemocyanin Revealed by Electron Cryomicroscopy.
Structure, 17, 2009
|
|
3IXW
 
 | | Scorpion Hemocyanin activated state pseudo atomic model built based on cryo-EM density map | | Descriptor: | Hemocyanin AA6 chain | | Authors: | Cong, Y, Zhang, Q, Woolford, D, Schweikardt, T, Khant, H, Ludtke, S, Chiu, W, Decker, H. | | Deposit date: | 2009-02-13 | | Release date: | 2009-06-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural Mechanism of SDS-Induced Enzyme Activity of Scorpion Hemocyanin Revealed by Electron Cryomicroscopy.
Structure, 17, 2009
|
|
3SQG
 
 | | Crystal structure of a methyl-coenzyme M reductase purified from Black Sea mats | | Descriptor: | (17[2]S)-17[2]-methylthio-coenzyme F43, 1-THIOETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Shima, S, Krueger, M, Weinert, T, Demmer, U, Thauer, R.K, Ermler, U. | | Deposit date: | 2011-07-05 | | Release date: | 2011-11-30 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a methyl-coenzyme M reductase from Black Sea mats that oxidize methane anaerobically.
Nature, 481, 2011
|
|
2B1P
 
 | | inhibitor complex of JNK3 | | Descriptor: | 3-{6-[(2-CHLOROPHENYL)AMINO]-1H-INDAZOL-3-YL}-5-{[4-(DIMETHYLAMINO)BUTANOYL]AMINO}BENZOIC ACID, BETA-MERCAPTOETHANOL, Mitogen-activated protein kinase 10, ... | | Authors: | Swahn, B.M, Huerta, F, Kallin, E, Malmstrom, J, Weigelt, T, Viklund, J, Womack, P, Xue, Y, Ohberg, L. | | Deposit date: | 2005-09-16 | | Release date: | 2006-09-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and synthesis of 6-anilinoindazoles as selective inhibitors of c-Jun N-terminal kinase-3
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2VD3
 
 | | The structure of histidine inhibited HisG from Methanobacterium thermoautotrophicum | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ATP PHOSPHORIBOSYLTRANSFERASE, ... | | Authors: | Lohkamp, B, Schweikert, T, Lapthorn, A.J. | | Deposit date: | 2007-09-28 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Structure of Histidine Inhibited Hisg from Methanobacterium Thermoautotrophicum
To be Published
|
|
5T89
 
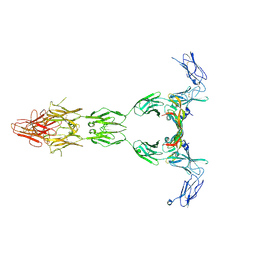 | | Crystal structure of VEGF-A in complex with VEGFR-1 domains D1-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Vascular endothelial growth factor A, ... | | Authors: | Markovic-Mueller, S, Stuttfeld, E, Asthana, M, Weinert, T, Bliven, S, Goldie, K.N, Kisko, K, Capitani, G, Ballmer-Hofer, K. | | Deposit date: | 2016-09-07 | | Release date: | 2017-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of the Full-length VEGFR-1 Extracellular Domain in Complex with VEGF-A.
Structure, 25, 2017
|
|
5NR4
 
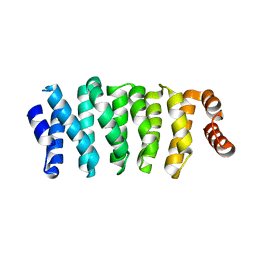 | | Crystal structure of Clasp2 TOG1 domain | | Descriptor: | CLIP-associating protein 2 | | Authors: | Sharma, A, Olieric, N, Weinert, T, Olieric, V, Steinmetz, M.O. | | Deposit date: | 2017-04-21 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.198 Å) | | Cite: | CLASP Suppresses Microtubule Catastrophes through a Single TOG Domain.
Dev. Cell, 46, 2018
|
|
8SS1
 
 | |
8SRZ
 
 | |
6GBV
 
 | |
6GAY
 
 | |
6GB3
 
 | |
6GBA
 
 | |
7XTQ
 
 | | Cryo-EM structure of the R399-bound GPBAR-Gs complex | | Descriptor: | G-protein coupled bile acid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ma, L, Yang, F, Wu, X, Mao, C, Sun, J, Yu, X, Zhang, Y, Zhang, P. | | Deposit date: | 2022-05-17 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis and molecular mechanism of biased GPBAR signaling in regulating NSCLC cell growth via YAP activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5GPO
 
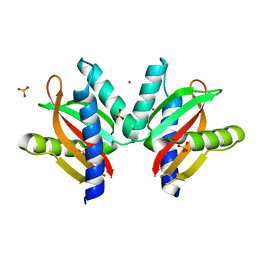 | | The sensor domain structure of the zinc-responsive histidine kinase CzcS from Pseudomonas Aeruginosa | | Descriptor: | SULFATE ION, Sensor protein CzcS, ZINC ION | | Authors: | Wang, D, Chen, W.Z, Huang, S.Q, Liu, X.C, Hu, Q.Y, Wei, T.B, Gan, J.H, Chen, H. | | Deposit date: | 2016-08-03 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural basis of Zn(II) induced metal detoxification and antibiotic resistance by histidine kinase CzcS in Pseudomonas aeruginosa
PLoS Pathog., 13, 2017
|
|
4DOY
 
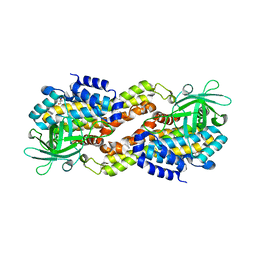 | | Crystal structure of Dibenzothiophene desulfurization enzyme C | | Descriptor: | Dibenzothiophene desulfurization enzyme C, GLYCEROL | | Authors: | Liu, S, Zhang, C, Zhu, D, Gu, L. | | Deposit date: | 2012-02-12 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Crystal structure of DszC from Rhodococcus sp. XP at 1.79 angstrom
Proteins, 82, 2014
|
|
4ZXA
 
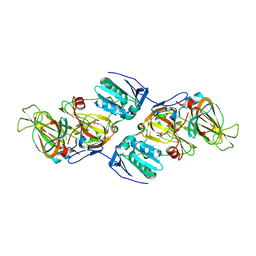 | | Crystal Structure of hydroquinone 1,2-dioxygenase PnpCD in complex with Cd2+ and 4-hydroxybenzonitrile | | Descriptor: | 4-hydroxybenzonitrile, CADMIUM ION, Hydroquinone dioxygenase large subunit, ... | | Authors: | Liu, S, Su, T, Zhang, C, Gu, L. | | Deposit date: | 2015-05-20 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.488 Å) | | Cite: | Crystal Structure of PnpCD, a Two-subunit Hydroquinone 1,2-Dioxygenase, Reveals a Novel Structural Class of Fe2+-dependent Dioxygenases.
J.Biol.Chem., 290, 2015
|
|
4ZXD
 
 | | Crystal Structure of hydroquinone 1,2-dioxygenase PnpCD | | Descriptor: | Hydroquinone dioxygenase large subunit, Hydroquinone dioxygenase small subunit | | Authors: | Liu, S, Su, T, Zhang, C, Gu, L. | | Deposit date: | 2015-05-20 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Crystal Structure of PnpCD, a Two-subunit Hydroquinone 1,2-Dioxygenase, Reveals a Novel Structural Class of Fe2+-dependent Dioxygenases.
J.Biol.Chem., 290, 2015
|
|
