6UP4
 
 | |
2N7H
 
 | | Hybrid structure of the Type 1 Pilus of Uropathogenic E.coli | | Descriptor: | FimA | | Authors: | Habenstein, B, Loquet, A, Giller, K, Vasa, S, Becker, S, Habeck, M, Lange, A. | | Deposit date: | 2015-09-11 | | Release date: | 2015-09-23 | | Last modified: | 2015-10-14 | | Method: | SOLID-STATE NMR | | Cite: | Hybrid Structure of the Type 1 Pilus of Uropathogenic Escherichia coli.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6ZGM
 
 | | Crystal Structure of the VIM-2 Acquired Metallo-beta-Lactamase in Complex with the thiazolecarboxylate inhibitor ANT2681 | | Descriptor: | 5-[[4-(carbamimidamidocarbamoylamino)-3,5-bis(fluoranyl)phenyl]sulfonylamino]-1,3-thiazole-4-carboxylic acid, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Docquier, J.D, Pozzi, C, Marcoccia, F, De Luca, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2020-06-19 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | ANT2681: SAR Studies Leading to the Identification of a Metallo-beta-lactamase Inhibitor with Potential for Clinical Use in Combination with Meropenem for the Treatment of Infections Caused by NDM-ProducingEnterobacteriaceae.
Acs Infect Dis., 6, 2020
|
|
2MME
 
 | | Hybrid structure of the Shigella flexneri MxiH Type three secretion system needle | | Descriptor: | MxiH | | Authors: | Demers, J.P, Habenstein, B, Loquet, A, Vasa, S.K, Becker, S, Baker, D, Lange, A, Sgourakis, N.G. | | Deposit date: | 2014-03-14 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.7 Å), SOLID-STATE NMR | | Cite: | High-resolution structure of the Shigella type-III secretion needle by solid-state NMR and cryo-electron microscopy.
Nat Commun, 5, 2014
|
|
6DMZ
 
 | | Solution structure of ZmD32 | | Descriptor: | Flower-specific gamma-thionin | | Authors: | Harvey, P.J, Craik, D.J. | | Deposit date: | 2018-06-05 | | Release date: | 2019-05-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Salt-Tolerant Antifungal and Antibacterial Activities of the Corn Defensin ZmD32.
Front Microbiol, 10, 2019
|
|
6ZXI
 
 | | Crystal Structure of the OXA-48 Carbapenem-Hydrolyzing Class D beta-Lactamase in Complex with the DBO inhibitor ANT3310 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CARBON DIOXIDE, ... | | Authors: | Docquier, J.D, Pozzi, C, De Luca, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2020-07-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of ANT3310 , a Novel Broad-Spectrum Serine beta-Lactamase Inhibitor of the Diazabicyclooctane Class, Which Strongly Potentiates Meropenem Activity against Carbapenem-Resistant Enterobacterales and Acinetobacter baumannii.
J.Med.Chem., 63, 2020
|
|
7JNN
 
 | |
7JN6
 
 | |
6XFP
 
 | | Crystal Structure of BRAF kinase domain bound to Belvarafenib | | Descriptor: | 4-amino-N-{1-[(3-chloro-2-fluorophenyl)amino]-6-methylisoquinolin-5-yl}thieno[3,2-d]pyrimidine-7-carboxamide, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Yin, J, Sudhamsu, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ARAF mutations confer resistance to the RAF inhibitor belvarafenib in melanoma.
Nature, 594, 2021
|
|
6MFP
 
 | | Crystal Structure of the RV305 C1-C2 specific ADCC potent antibody DH677.3 Fab in complex with HIV-1 clade A/E gp120 and M48U1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Tolbert, W.D, Young, B, Pazgier, M. | | Deposit date: | 2018-09-11 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Boosting with AIDSVAX B/E Enhances Env Constant Region 1 and 2 Antibody-Dependent Cellular Cytotoxicity Breadth and Potency.
J.Virol., 94, 2020
|
|
6VBP
 
 | |
6MFJ
 
 | |
6VBQ
 
 | |
6VBO
 
 | |
8FHY
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with neutralizing antibody WRAIR-5021 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MALONATE ION, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2022-12-15 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Diverse array of neutralizing antibodies elicited upon Spike Ferritin Nanoparticle vaccination in rhesus macaques.
Nat Commun, 15, 2024
|
|
8FI9
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody WRAIR-5001 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2022-12-15 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Diverse array of neutralizing antibodies elicited upon Spike Ferritin Nanoparticle vaccination in rhesus macaques.
Nat Commun, 15, 2024
|
|
5WKH
 
 | | D30 TCR in complex with HLA-A*11:01-GTS3 | | Descriptor: | Beta-2-microglobulin, D30 TCR beta chain, GTS3 peptide, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-25 | | Release date: | 2017-09-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8(+) T cell response.
Nat. Immunol., 18, 2017
|
|
5WKF
 
 | | D30 TCR in complex with HLA-A*11:01-GTS1 | | Descriptor: | Beta-2-microglobulin, D30 TCR beta chain, GTS1 peptide, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-25 | | Release date: | 2017-09-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8(+) T cell response.
Nat. Immunol., 18, 2017
|
|
5WJL
 
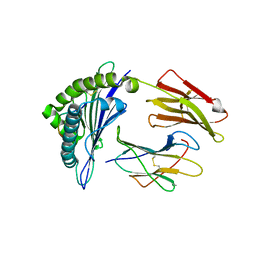 | | Crystal Structure of HLA-A*11:01 with GTS1 peptide | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GTS1 peptide, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-23 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8(+) T cell response.
Nat. Immunol., 18, 2017
|
|
5WJN
 
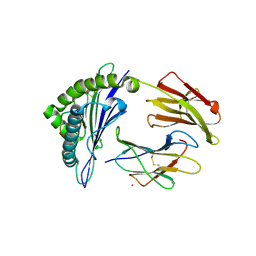 | | Crystal Structure of HLA-A*11:01-GTS3 | | Descriptor: | Beta-2-microglobulin, GTS3 peptide, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-23 | | Release date: | 2017-09-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8(+) T cell response.
Nat. Immunol., 18, 2017
|
|
5UIX
 
 | |
1PRO
 
 | | HIV-1 PROTEASE DIMER COMPLEXED WITH A-98881 | | Descriptor: | (5R,6R)-2,4-BIS-(4-HYDROXY-3-METHOXYBENZYL)-1,5-DIBENZYL-3-OXO-6-HYDROXY-1,2,4-TRIAZACYCLOHEPTANE, HIV-1 PROTEASE | | Authors: | Park, C.H, Kong, X.P, Dealwis, C.G. | | Deposit date: | 1995-07-18 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel, picomolar inhibitor of human immunodeficiency virus type 1 protease.
J.Med.Chem., 39, 1996
|
|
8F5U
 
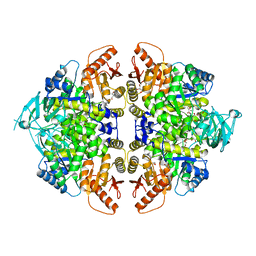 | | Rabbit muscle pyruvate kinase in complex with magnesium, potassium and pyruvate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Holyoak, T, Fenton, A.W. | | Deposit date: | 2022-11-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | PYK-SubstitutionOME: an integrated database containing allosteric coupling, ligand affinity and mutational, structural, pathological, bioinformatic and computational information about pyruvate kinase isozymes.
Database (Oxford), 2023, 2023
|
|
8F5T
 
 | | Rabbit muscle pyruvate kinase in complex with sodium and magnesium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Holyoak, T, Fenton, A.W. | | Deposit date: | 2022-11-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | PYK-SubstitutionOME: an integrated database containing allosteric coupling, ligand affinity and mutational, structural, pathological, bioinformatic and computational information about pyruvate kinase isozymes.
Database (Oxford), 2023, 2023
|
|
8F6M
 
 | | Complex of Rabbit muscle pyruvate kinase with ADP and the phosphonate analogue of PEP mimicking the Michaelis complex. | | Descriptor: | (E)-2-METHYL-3-PHOSPHONOACRYLATE, ADENOSINE-5'-DIPHOSPHATE, ALANINE, ... | | Authors: | Holyoak, T, Fenton, A.W. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | PYK-SubstitutionOME: an integrated database containing allosteric coupling, ligand affinity and mutational, structural, pathological, bioinformatic and computational information about pyruvate kinase isozymes.
Database (Oxford), 2023, 2023
|
|
