8JYP
 
 | | Structure of SARS-CoV-2 XBB.1.5 spike RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYN
 
 | | Structure of SARS-CoV-2 XBB.1.5 spike glycoprotein in complex with ACE2 (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYO
 
 | | Structure of SARS-CoV-2 XBB.1.5 spike glycoprotein in complex with ACE2 (2-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
5Y6Q
 
 | |
6CBY
 
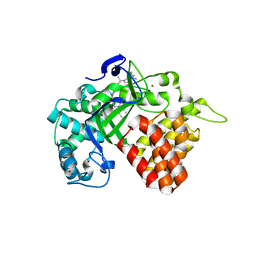 | | Crystal structure of human SET and MYND Domain Containing protein 2 with MTF9975 | | Descriptor: | N-lysine methyltransferase SMYD2, ZINC ION, [3-(4-amino-6-methyl-1H-imidazo[4,5-c]pyridin-1-yl)-3-methylazetidin-1-yl][1-({1-[(1R)-cyclohept-2-en-1-yl]piperidin-4-yl}methyl)-1H-pyrrol-3-yl]methanone | | Authors: | ZENG, H, DONG, A, Hutchinson, A, Seitova, A, TATLOCK, J, KUMPF, R, OWEN, A, TAYLOR, A, Casimiro-Garcia, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-05 | | Release date: | 2018-03-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Selective, Small-Molecule Co-Factor Binding Site Inhibition of a Su(var)3-9, Enhancer of Zeste, Trithorax Domain Containing Lysine Methyltransferase.
J.Med.Chem., 62, 2019
|
|
5VYG
 
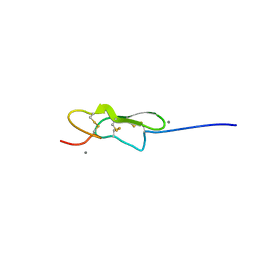 | | Crystal structure of hFA9 EGF repeat with O-glucose trisaccharide | | Descriptor: | CALCIUM ION, Coagulation factor IX, alpha-D-xylopyranose-(1-3)-alpha-D-xylopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Yu, H.J, Li, H.L. | | Deposit date: | 2017-05-25 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | O-Glycosylation modulates the stability of epidermal growth factor-like repeats and thereby regulates Notch trafficking
J. Biol. Chem., 292, 2017
|
|
6P7I
 
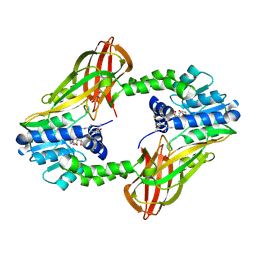 | | Crystal structure of Human PRMT6 in complex with S-Adenosyl-L-Homocysteine and YS17-117 Compound | | Descriptor: | GLYCEROL, N-[3-(4-{[(2-aminoethyl)(methyl)amino]methyl}-1H-pyrrol-3-yl)phenyl]prop-2-enamide, N-[3-(4-{[(2-aminoethyl)(methyl)amino]methyl}-1H-pyrrol-3-yl)phenyl]propanamide, ... | | Authors: | Halabelian, L, Dong, A, Zeng, H, Li, Y, Seitova, A, Hutchinson, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-05 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a First-in-Class Protein Arginine Methyltransferase 6 (PRMT6) Covalent Inhibitor
J.Med.Chem., 63, 2020
|
|
8IF2
 
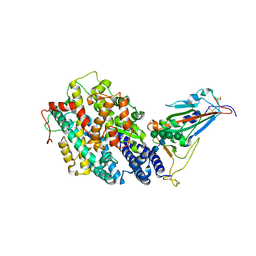 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BQ.1.1 variant spike protein in complex with its receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Kimura, K, Suzuki, T, Hashiguchi, T. | | Deposit date: | 2023-02-17 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Convergent evolution of SARS-CoV-2 Omicron subvariants leading to the emergence of BQ.1.1 variant.
Nat Commun, 14, 2023
|
|
6D2L
 
 | | Crystal structure of human CARM1 with (S)-SKI-72 | | Descriptor: | (2S,5S)-2-amino-6-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-5-[(benzylamino)methyl]-N-[2-(4-hydroxyphenyl)ethyl]hexanamide, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | DONG, A, ZENG, H, WALKER, J.R, Hutchinson, A, Seitova, A, LUO, M, CAI, X.C, KE, W, WANG, J, SHI, C, ZHENG, W, LEE, J.P, IBANEZ, G, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-13 | | Release date: | 2018-05-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A chemical probe of CARM1 alters epigenetic plasticity against breast cancer cell invasion.
Elife, 8, 2019
|
|
4YZI
 
 | | Crystal structure of blue-shifted channelrhodopsin mutant (T198G/G202A) | | Descriptor: | OLEIC ACID, RETINAL, Sensory opsin A,Archaeal-type opsin 2, ... | | Authors: | Kato, H.E, Kamiya, M, Ishitani, R, Hayashi, S, Nureki, O. | | Deposit date: | 2015-03-25 | | Release date: | 2015-05-27 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomistic design of microbial opsin-based blue-shifted optogenetics tools.
Nat Commun, 6, 2015
|
|
6IZZ
 
 | |
6J00
 
 | |
6AAJ
 
 | | Crystal structure of JAK2 in complex with peficitinib | | Descriptor: | 4-[[(1S,3R)-5-oxidanyl-2-adamantyl]amino]-1H-pyrrolo[2,3-b]pyridine-5-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Amano, Y, Tateishi, Y. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-15 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery and structural characterization of peficitinib (ASP015K) as a novel and potent JAK inhibitor
Bioorg. Med. Chem., 26, 2018
|
|
6AAM
 
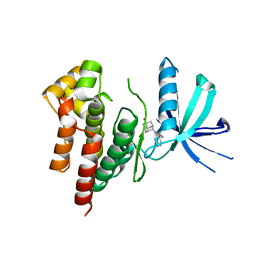 | | Crystal structure of TYK2 in complex with peficitinib | | Descriptor: | 4-[[(1S,3R)-5-oxidanyl-2-adamantyl]amino]-1H-pyrrolo[2,3-b]pyridine-5-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Nomura, N, Tomimoto, Y. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery and structural characterization of peficitinib (ASP015K) as a novel and potent JAK inhibitor
Bioorg. Med. Chem., 26, 2018
|
|
6AAH
 
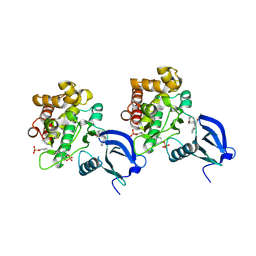 | | Crystal structure of JAK1 in complex with peficitinib | | Descriptor: | 4-[[(1S,3R)-5-oxidanyl-2-adamantyl]amino]-1H-pyrrolo[2,3-b]pyridine-5-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Amano, Y. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-15 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery and structural characterization of peficitinib (ASP015K) as a novel and potent JAK inhibitor
Bioorg. Med. Chem., 26, 2018
|
|
6AAK
 
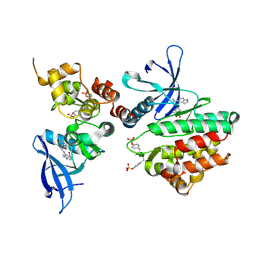 | | Crystal structure of JAK3 in complex with peficitinib | | Descriptor: | 4-[[(1S,3R)-5-oxidanyl-2-adamantyl]amino]-1H-pyrrolo[2,3-b]pyridine-5-carboxamide, Tyrosine-protein kinase JAK3 | | Authors: | Amano, Y. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-15 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery and structural characterization of peficitinib (ASP015K) as a novel and potent JAK inhibitor
Bioorg. Med. Chem., 26, 2018
|
|
3AQA
 
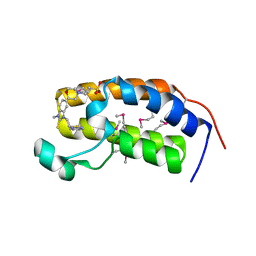 | | Crystal structure of the human BRD2 BD1 bromodomain in complex with a BRD2-interactive compound, BIC1 | | Descriptor: | 1-[2-(1H-benzimidazol-2-ylsulfanyl)ethyl]-3-methyl-1,3-dihydro-2H-benzimidazole-2-thione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2 | | Authors: | Umehara, T, Nakamura, Y, Terada, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-10-27 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Real-Time Imaging of Histone H4K12-Specific Acetylation Determines the Modes of Action of Histone Deacetylase and Bromodomain Inhibitors
Chem.Biol., 18, 2011
|
|
8I5I
 
 | |
8I5H
 
 | |
7SSE
 
 | | Crystal structure of the WDR domain of human DCAF1 in complex with CYCA-117-70 | | Descriptor: | DDB1- and CUL4-associated factor 1, N-[(3R)-1-(3-fluorophenyl)piperidin-3-yl]-6-(morpholin-4-yl)pyrimidin-4-amine | | Authors: | Kimani, S, Owen, J, Li, A, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Shahani, V.M, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery of a Novel DCAF1 Ligand Using a Drug-Target Interaction Prediction Model: Generalizing Machine Learning to New Drug Targets.
J.Chem.Inf.Model., 63, 2023
|
|
1VEA
 
 | | Crystal Structure of HutP, an RNA binding antitermination protein | | Descriptor: | Hut operon positive regulatory protein, N-(2-NAPHTHYL)HISTIDINAMIDE | | Authors: | Kumarevel, T.S, Fujimoto, Z, Karthe, P, Oda, M, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2004-03-29 | | Release date: | 2004-07-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Activated HutP; An RNA Binding Protein that Regulates Transcription of the hut Operon in Bacillus subtilis
Structure, 12, 2004
|
|
8IOS
 
 | | Structure of the SARS-CoV-2 XBB.1 spike glycoprotein (closed-1 state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8IOU
 
 | | Structure of SARS-CoV-2 XBB.1 spike glycoprotein in complex with ACE2 (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8IOV
 
 | | Structure of SARS-CoV-2 XBB.1 spike RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8IOT
 
 | | Structure of the SARS-CoV-2 XBB.1 spike glycoprotein (closed-2 state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
