6WQL
 
 | |
6WQJ
 
 | |
6XK9
 
 | | Cereblon in complex with DDB1, CC-90009, and GSPT1 | | Descriptor: | 2-(4-chlorophenyl)-N-({2-[(3S)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-5-yl}methyl)-2,2-difluoroacetamide, DNA damage-binding protein 1, Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, ... | | Authors: | Clayton, T.L, Tran, E.T, Zhu, J, Pagarigan, B.E, Matyskiela, M.E, Chamberlain, P.P. | | Deposit date: | 2020-06-25 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | CC-90009, a novel cereblon E3 ligase modulator, targets acute myeloid leukemia blasts and leukemia stem cells.
Blood, 137, 2021
|
|
6O44
 
 | | Insight into subtilisin E-S7 cleavage pattern based on crystal structure and hydrolysates peptide analysis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Nattokinase, ... | | Authors: | Tang, H, Shi, K, Aihara, H. | | Deposit date: | 2019-02-28 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Insight into subtilisin E-S7 cleavage pattern based on crystal structure and hydrolysates peptide analysis.
Biochem. Biophys. Res. Commun., 512, 2019
|
|
7OOM
 
 | |
6XB9
 
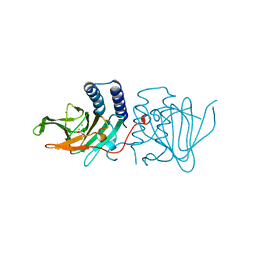 | | Crystal structure of Azotobacter vinelandii 3-mercaptopropionic acid dioxygenase in complex with 3-hydroxypropionic acid | | Descriptor: | 3-HYDROXY-PROPANOIC ACID, CHLORIDE ION, Cysteine dioxygenase type I protein, ... | | Authors: | Kiser, P.D, Khadka, N, Shi, W, Pierce, B.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of 3-mercaptopropionic acid dioxygenase with a substrate analog reveals bidentate substrate binding at the iron center.
J.Biol.Chem., 296, 2021
|
|
3F0G
 
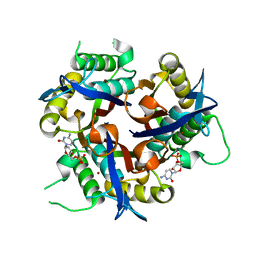 | |
6EAC
 
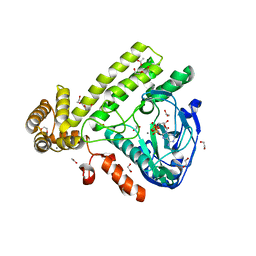 | | Pseudomonas syringae SelO | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Tomchick, D.R, Tagliabracci, V.S, Sreelatha, A. | | Deposit date: | 2018-08-02 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Protein AMPylation by an Evolutionarily Conserved Pseudokinase.
Cell, 175, 2018
|
|
8OYP
 
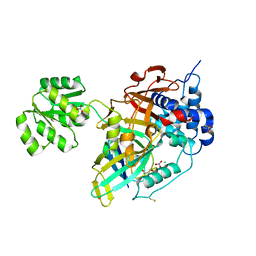 | | Crystal structure of Ubiquitin specific protease 11 (USP11) in complex with a substrate mimetic | | Descriptor: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Maurer, S.K, Caulton, S.G, Ward, S.J, Emsley, J, Dreveny, I. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Ubiquitin-specific protease 11 structure in complex with an engineered substrate mimetic reveals a molecular feature for deubiquitination selectivity.
J.Biol.Chem., 299, 2023
|
|
6VCG
 
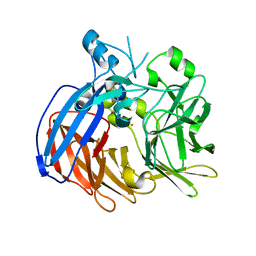 | | Crystal structure of Nitrosotalea devanaterra carotenoid cleavage dioxygenase, cobalt form | | Descriptor: | CHLORIDE ION, COBALT (II) ION, SODIUM ION, ... | | Authors: | Daruwalla, A, Shi, W, Kiser, P.D. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PBT
 
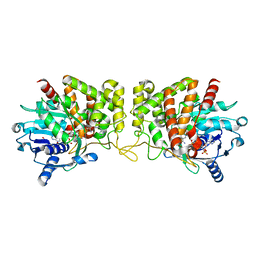 | | Pseudopaline Dehydrogenase with (R)-Pseudopaline Soaked 2 hours | | Descriptor: | 1,2-ETHANEDIOL, N-[(3S)-3-amino-3-carboxypropyl]-L-histidine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | McFarlane, J.S, Lamb, A.L. | | Deposit date: | 2019-06-14 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Staphylopine and pseudopaline dehydrogenase from bacterial pathogens catalyze reversible reactions and produce stereospecific metallophores.
J.Biol.Chem., 294, 2019
|
|
5UDD
 
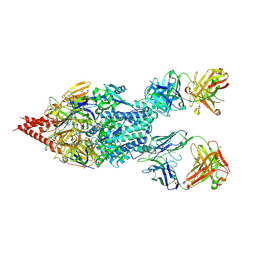 | | Crystal Structure of RSV F B9320 Bound to MEDI8897 | | Descriptor: | Fusion glycoprotein F0, MEDI8897 Fab Heavy Chain, MEDI8897 Fab Light Chain, ... | | Authors: | McLellan, J.S. | | Deposit date: | 2016-12-26 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | A highly potent extended half-life antibody as a potential RSV vaccine surrogate for all infants.
Sci Transl Med, 9, 2017
|
|
6PBN
 
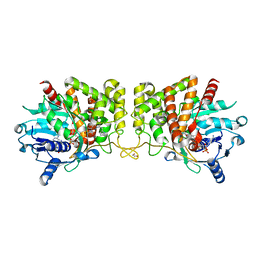 | | Pseudopaline Dehydrogenase with (R)-Pseudopaline Soaked 1 hour | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, N-[(3S)-3-amino-3-carboxypropyl]-L-histidine, ... | | Authors: | McFarlane, J.S, Lamb, A.L. | | Deposit date: | 2019-06-14 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Staphylopine and pseudopaline dehydrogenase from bacterial pathogens catalyze reversible reactions and produce stereospecific metallophores.
J.Biol.Chem., 294, 2019
|
|
6B8U
 
 | | Crystals Structure of B-Raf kinase domain in complex with an Imidazopyridinyl benzamide inhibitor | | Descriptor: | Serine/threonine-protein kinase B-raf, ~{N}-[3-(2-acetamidoimidazo[1,2-a]pyridin-6-yl)-4-methyl-phenyl]-3-(trifluoromethyl)benzamide | | Authors: | Appleton, B.A, Murray, J, Shafer, C.M. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Imidazo[1,2-a]pyridin-6-yl-benzamide analogs as potent RAF inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6VCF
 
 | | Crystal structure of Nitrosotalea devanaterra carotenoid cleavage dioxygenase, iron form | | Descriptor: | BICARBONATE ION, CHLORIDE ION, FE (II) ION, ... | | Authors: | Daruwalla, A, Shi, W, Kiser, P.D. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.687 Å) | | Cite: | Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VHS
 
 | | Crystal structure of CTX-M-14 in complex with beta-lactamase inhibitor ETX1317 | | Descriptor: | (2R)-({[(3R,6S)-6-carbamoyl-1-formyl-4-methyl-1,2,3,6-tetrahydropyridin-3-yl]amino}oxy)(fluoro)acetic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Sacco, M.D, Chen, Y. | | Deposit date: | 2020-01-10 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Discovery of an Orally Available Diazabicyclooctane Inhibitor (ETX0282) of Class A, C, and D Serine beta-Lactamases.
J.Med.Chem., 63, 2020
|
|
6PBM
 
 | | Pseudopaline Dehydrogenase with NADP+ bound | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pseudopaline Dehydrogenase | | Authors: | McFarlane, J.S, Lamb, A.L. | | Deposit date: | 2019-06-14 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Staphylopine and pseudopaline dehydrogenase from bacterial pathogens catalyze reversible reactions and produce stereospecific metallophores.
J.Biol.Chem., 294, 2019
|
|
5ED3
 
 | | crystal structure of human Hint1 complexing with AP5A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2015-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.309 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
6VQF
 
 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C (NHIS-RORGT(244- 487)-L6-SRC1(678-692)) IN COMPLEX WITH AN INVERSE AGONIST | | Descriptor: | (1R,3S,4R)-4-[(3aR,9bR)-9b-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-1,2,3a,4,5,9b-hexahydro-3H-benzo[e]indole-3-carbonyl]-3-methylcyclohexane-1-carboxylic acid, GLYCEROL, Nuclear receptor ROR-gamma | | Authors: | Sack, J.S. | | Deposit date: | 2020-02-05 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of BMS-986251: A Clinically Viable, Potent, and Selective ROR gamma t Inverse Agonist.
Acs Med.Chem.Lett., 11, 2020
|
|
6PAK
 
 | | Insight into subtilisin E-S7 cleavage pattern based on crystal structure and hydrolysates peptide analysis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Subtilisin E | | Authors: | Tang, H, Shi, K, Aihara, H. | | Deposit date: | 2019-06-11 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Enhancing subtilisin thermostability through a modified normalized B-factor analysis and loop-grafting strategy.
J.Biol.Chem., 294, 2019
|
|
6PDX
 
 | |
6PBP
 
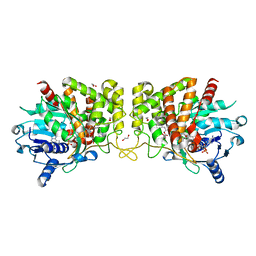 | | Pseudopaline Dehydrogenase with (S)-Pseudopaline Soaked 1 hour | | Descriptor: | 1,2-ETHANEDIOL, N-[(1S)-1-carboxy-3-{[(1S)-1-carboxy-2-(1H-imidazol-5-yl)ethyl]amino}propyl]-L-glutamic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | McFarlane, J.S, Lamb, A.L. | | Deposit date: | 2019-06-14 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Staphylopine and pseudopaline dehydrogenase from bacterial pathogens catalyze reversible reactions and produce stereospecific metallophores.
J.Biol.Chem., 294, 2019
|
|
8A6O
 
 | |
8A6S
 
 | |
8A6N
 
 | |
