6GYH
 
 | | Crystal structure of the light-driven proton pump Coccomyxa subellipsoidea Rhodopsin CsR | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Family A G protein-coupled receptor-like protein, ... | | Authors: | Szczepek, M, Schmidt, A, Scheerer, P. | | Deposit date: | 2018-06-29 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of a light-gated proton channel based on the crystal structure ofCoccomyxarhodopsin.
Sci.Signal., 12, 2019
|
|
2B98
 
 | | Crystal Structure of an archaeal pentameric riboflavin synthase | | Descriptor: | Riboflavin synthase | | Authors: | Ramsperger, A, Augustin, M, Schott, A.K, Gerhardt, S, Krojer, T, Eisenreich, W, Illarionov, B, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2005-10-11 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of an Archaeal Pentameric Riboflavin Synthase in Complex with a Substrate Analog Inhibitor: stereochemical implications
J.Biol.Chem., 281, 2006
|
|
5DL7
 
 | |
5DQ4
 
 | |
4KNB
 
 | | C-Met in complex with OSI ligand | | Descriptor: | 7-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]furo[3,2-c]pyridin-6-amine, GAMMA-BUTYROLACTONE, Hepatocyte growth factor receptor | | Authors: | Wang, J, Steinig, A.G, Li, A.H, Chen, X, Dong, H, Ferraro, C, Jin, M, Kadalbajoo, M, Kleinberg, A, Stolz, K.M, Tavares-Greco, P.A, Wang, T, Albertella, M.R, Peng, Y, Crew, L, Kahler, J. | | Deposit date: | 2013-05-09 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel 6-aminofuro[3,2-c]pyridines as potent, orally efficacious inhibitors of cMET and RON kinases.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1QLR
 
 | | CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF A HUMAN MONOCLONAL IgM COLD AGGLUTININ | | Descriptor: | IGM FAB REGION IV-J(H4)-C (KAU COLD AGGLUTININ), IGM KAPPA CHAIN V-III (KAU COLD AGGLUTININ), alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Carvalho, J.G, Cauerhff, A, Goldbaum, F, Leoni, J, Polikarpov, I. | | Deposit date: | 1999-09-11 | | Release date: | 2000-09-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Three-dimensional structure of the Fab from a human IgM cold agglutinin.
J Immunol., 165, 2000
|
|
2A8B
 
 | | Crystal Structure of the Catalytic Domain of Human Tyrosine Phosphatase Receptor, Type R | | Descriptor: | CHLORIDE ION, Receptor-type tyrosine-protein phosphatase R | | Authors: | Ugochukwu, E, Eswaran, J, Barr, A, Longman, E, Arrowsmith, C, Edwards, A, Sundstrom, M, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-07-07 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures and inhibitor identification for PTPN5, PTPRR and PTPN7: a family of human MAPK-specific protein tyrosine phosphatases.
Biochem.J., 395, 2006
|
|
2KVY
 
 | | NMR solution structure of the 4:1 complex between an uncharged distamycin A analogue and [d(TGGGGT)]4 | | Descriptor: | 4-amino-1-methyl-N-{1-methyl-5-[(1-methyl-5-{[3-(methylamino)-3-oxopropyl]carbamoyl}-1H-pyrrol-3-yl)carbamoyl]-1H-pyrrol-3-yl}-1H-pyrrole-2-carboxamide, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3') | | Authors: | Cosconati, S, Marinelli, L, Trotta, R, Virno, A, De Tito, S, Romagnoli, R, Pagano, B, Limongelli, V, Giancola, C, Baraldi, P, Mayol, L, Novellino, E, Randazzo, A. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and conformational requisites in DNA quadruplex groove binding: another piece to the puzzle.
J.Am.Chem.Soc., 132, 2010
|
|
6H0N
 
 | |
3T7V
 
 | | Crystal structure of methylornithine synthase (PylB) | | Descriptor: | 5-amino-D-isoleucine, IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE, ... | | Authors: | Quitterer, F, List, A, Eisenreich, W, Bacher, A, Groll, M. | | Deposit date: | 2011-07-31 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of methylornithine synthase (PylB): insights into the pyrrolysine biosynthesis.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3DYF
 
 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-461 and Isopentyl Diphosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-FLUORO-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM, ACETATE ION, ... | | Authors: | Cao, R, Gao, Y, Robinson, H, Goddard, A, Oldfield, E. | | Deposit date: | 2008-07-27 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Lipophilic bisphosphonates as dual farnesyl/geranylgeranyl diphosphate synthase inhibitors: an X-ray and NMR investigation.
J.Am.Chem.Soc., 131, 2009
|
|
2ZMN
 
 | | Crystal Structure of basic winged bean lectin in complex with Gal-alpha- 1,6 Glc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Basic agglutinin, CALCIUM ION, ... | | Authors: | Kulkarni, K.A, Katiyar, S, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2008-04-19 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and sugar-specificity of basic winged-bean lectin: structures of new disaccharide complexes and a comparative study with other known disaccharide complexes of the lectin.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2CGY
 
 | | STRUCTURE OF HELIX POMATIA AGGLUTININ WITH FORSMANN ANTIGEN | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, AGGLUTININ | | Authors: | Sanchez, J.-F, Lescar, J, Audfray, A, Gautier, C, Chazalet, V, Gagnon, J, Breton, C, Imberty, A, Mitchell, E.P. | | Deposit date: | 2006-03-10 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Recognition of Breast and Colon Cancer Epitopes Tn Antigen and Forssman Disaccharide by Helix Pomatia Lectin.
Glycobiology, 17, 2007
|
|
2CGZ
 
 | | Structure of Helix Pomatia agglutinin with Tn antigen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, AGGLUTININ, ... | | Authors: | Sanchez, J.-F, Lescar, J, Audfray, A, Gautier, C, Chazalet, V, Gagnon, J, Breton, C, Imberty, A, Mitchell, E.P. | | Deposit date: | 2006-03-10 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Recognition of Breast and Colon Cancer Epitopes Tn Antigen and Forssman Disaccharide by Helix Pomatia Lectin.
Glycobiology, 17, 2007
|
|
2KZ4
 
 | | Solution structure of protein SF1141 from Shigella flexneri 2a, Northeast structural genomics consortium (NESG) target SFT2 | | Descriptor: | Putative head-tail adaptor | | Authors: | Lemak, A, Yee, A, Garcia, M, Lee, H.-W, Prestegard, J, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-11 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein SF1141 from Shigella flexneri 2a
To be Published
|
|
2ZKN
 
 | |
4KL7
 
 | | Crystal structure of the catalytic domain of RpfB from Mycobacterium tuberculosis | | Descriptor: | Resuscitation-promoting factor RpfB, SULFATE ION | | Authors: | Squeglia, F, Romano, M, Ruggiero, A, Berisio, R. | | Deposit date: | 2013-05-07 | | Release date: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Carbohydrate Recognition by RpfB from Mycobacterium tuberculosis Unveiled by Crystallographic and Molecular Dynamics Analyses.
Biophys.J., 104, 2013
|
|
3H5S
 
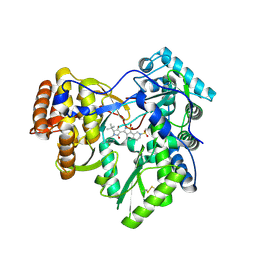 | | Hepatitis C virus polymerase NS5B with saccharin inhibitor | | Descriptor: | (5S)-5-tert-butyl-1-(4-fluoro-3-methylbenzyl)-4-hydroxy-3-[8-(methylsulfonyl)-1,1-dioxido-6,7,8,9-tetrahydroisothiazolo[4,5-h]isoquinolin-3-yl]-1,5-dihydro-2H-pyrrol-2-one, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Wong, A. | | Deposit date: | 2009-04-22 | | Release date: | 2009-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-nucleoside inhibitors of HCV polymerase NS5B. Part 4: structure-based design, synthesis, and biological evaluation of benzo[d]isothiazole-1,1-dioxides
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3GUD
 
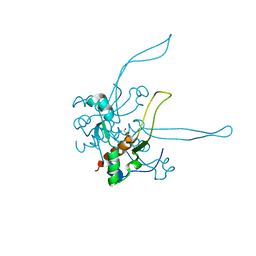 | | Crystal structure of a novel intramolecular chaperon | | Descriptor: | BROMIDE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-29 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an intramolecular chaperone mediating triple-beta-helix folding.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2CMR
 
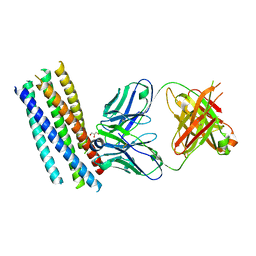 | | Crystal structure of the HIV-1 neutralizing antibody D5 Fab bound to the gp41 inner-core mimetic 5-helix | | Descriptor: | D5, GLYCEROL, TRANSMEMBRANE GLYCOPROTEIN | | Authors: | Luftig, M.A, Mattu, M, Di Giovine, P, Geleziunas, R, Hrin, R, Barbato, G, Bianchi, E, Miller, M.D, Pessi, A, Carfi, A. | | Deposit date: | 2006-05-11 | | Release date: | 2006-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for HIV-1 Neutralization by a Gp41 Fusion Intermediate-Directed Antibody
Nat.Struct.Mol.Biol., 13, 2006
|
|
3GUT
 
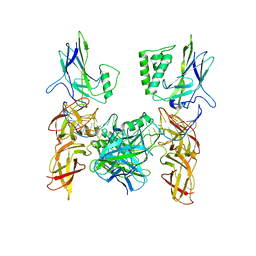 | | Crystal structure of a higher-order complex of p50:RelA bound to the HIV-1 LTR | | Descriptor: | HIV-LTR Core Forward Strand, HIV-LTR Core Reverse Strand, Nuclear factor NF-kappa-B p105 subunit, ... | | Authors: | Stroud, J.C, Oltman, A.J, Han, A, Bates, D.L, Chen, L. | | Deposit date: | 2009-03-30 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural basis of HIV-1 activation by NF-kappaB--a higher-order complex of p50:RelA bound to the HIV-1 LTR.
J.Mol.Biol., 393, 2009
|
|
2K5F
 
 | | Solution NMR structure of FeoA protein from Chlorobium tepidum. Northeast Structural Genomics Consortium target CtR121 | | Descriptor: | Ferrous iron transport protein A | | Authors: | Eletsky, A, Sathyamoorthy, B, Mills, J.L, Zeri, A, Zhao, L, Hamilton, K, Foote, E.L, Xiao, R, Nair, R, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-27 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of FeoA protein from Chlorobium tepidum. Northeast Structural Genomics Consortium target CtR121
To be Published
|
|
1B37
 
 | | A 30 ANGSTROM U-SHAPED CATALYTIC TUNNEL IN THE CRYSTAL STRUCTURE OF POLYAMINE OXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (POLYAMINE OXIDASE), ... | | Authors: | Binda, C, Coda, A, Angelini, R, Federico, R, Ascenzi, P, Mattevi, A. | | Deposit date: | 1998-12-17 | | Release date: | 1999-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 30-angstrom-long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase.
Structure Fold.Des., 7, 1999
|
|
1IH5
 
 | | CRYSTAL STRUCTURE OF AQUAPORIN-1 | | Descriptor: | AQUAPORIN-1 | | Authors: | Ren, G, Reddy, V.S, Cheng, A, Melnyk, P, Mitra, A.K. | | Deposit date: | 2001-04-18 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.7 Å) | | Cite: | Visualization of a water-selective pore by electron crystallography in vitreous ice.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2KVR
 
 | | Solution NMR structure of human ubiquitin specific protease Usp7 UBL domain (residues 537-664). NESG target hr4395c/ SGC-Toronto | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Bezsonova, I, Lemak, A, Avvakumov, G, Xue, S, Dhe-Paganon, S, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-25 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | solution structure of the ubiquitin specific protease Usp7 ubiquitin-like domain
To be Published
|
|
