7L9L
 
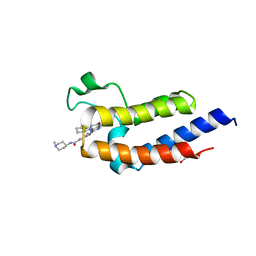 | | Crystal structure of the second bromodomain (BD2) of human BRD3 bound to BI2536 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 3 | | Authors: | Karim, M.R, Bikowitz, M.J, Schonbrunn, E. | | Deposit date: | 2021-01-04 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7L9K
 
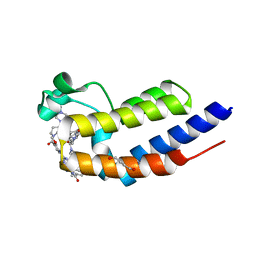 | | Crystal structure of the second bromodomain (BD2) of human BRD2 bound to LRRK2-IN-1 | | Descriptor: | 1,4-BUTANEDIOL, 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 2 | | Authors: | Karim, M.R, Bikowitz, M.J, Schonbrunn, E. | | Deposit date: | 2021-01-04 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LB4
 
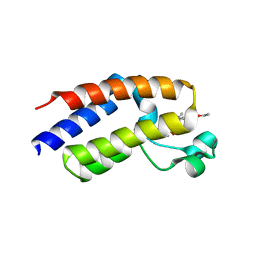 | |
7L9J
 
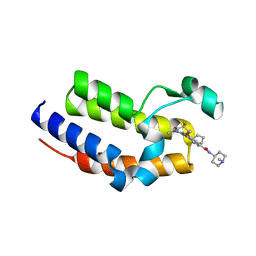 | | Crystal structure of the second bromodomain (BD2) of human BRD2 bound to Ro3280 | | Descriptor: | 4-[(9-cyclopentyl-7,7-difluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl)amino]-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 2, CHLORIDE ION | | Authors: | Karim, M.R, Bikowitz, M.J, Schonbrunn, E. | | Deposit date: | 2021-01-04 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LAZ
 
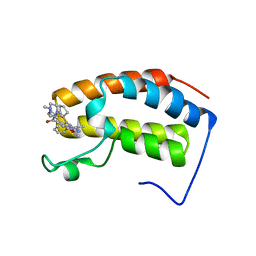 | | Crystal structure of the first bromodomain (BD1) of human BRD3 bound to ERK5-IN-1 | | Descriptor: | 11-cyclopentyl-2-({2-ethoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 3 | | Authors: | Karim, M.R, Bikowitz, M.J, Schonbrunn, E. | | Deposit date: | 2021-01-07 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LBT
 
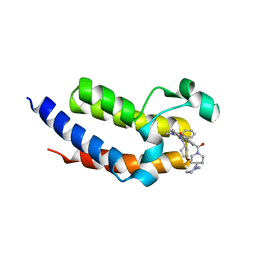 | | Crystal structure of the second bromodomain (BD2) of human BRD3 bound to ERK5-IN-1 | | Descriptor: | 11-cyclopentyl-2-({2-ethoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 3 | | Authors: | Karim, M.R, Bikowitz, M.J, Schonbrunn, E. | | Deposit date: | 2021-01-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LAY
 
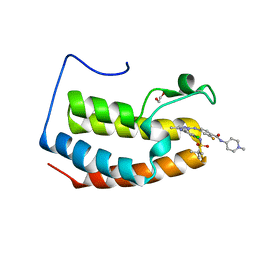 | | Crystal structure of the first bromodomain (BD1) of human BRD3 bound to SG3-179 | | Descriptor: | 1,2-ETHANEDIOL, 4-[[4-[[3-(~{tert}-butylsulfonylamino)-4-chloranyl-phenyl]amino]-5-methyl-pyrimidin-2-yl]amino]-2-fluoranyl-~{N}-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 3, ... | | Authors: | Karim, M.R, Bikowitz, M.J, Schonbrunn, E. | | Deposit date: | 2021-01-07 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
5UJX
 
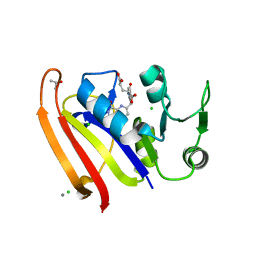 | | Crystal structure of DHFR in 20% Isopropanol | | Descriptor: | CALCIUM ION, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Cuneo, M.J, Agarwal, P.K. | | Deposit date: | 2017-01-19 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulating Enzyme Activity by Altering Protein Dynamics with Solvent.
Biochemistry, 57, 2018
|
|
4PY4
 
 | |
2F8B
 
 | |
2R5L
 
 | | Crystal structure of lactoperoxidase at 2.4A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Singh, A.K, Singh, N, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Lactoperoxidase at 2.4 A Resolution.
J.Mol.Biol., 376, 2007
|
|
2DT0
 
 | | Crystal structure of the complex of goat signalling protein with the trimer of N-acetylglucosamine at 2.45A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kumar, J, Ethayathulla, A.S, Srivastava, D.B, Singh, N, Sharma, S, Bhushan, A, Srinivasan, A, Singh, T.P. | | Deposit date: | 2006-07-09 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Carbohydrate-binding properties of goat secretory glycoprotein (SPG-40) and its functional implications: structures of the native glycoprotein and its four complexes with chitin-like oligosaccharides
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
1T1O
 
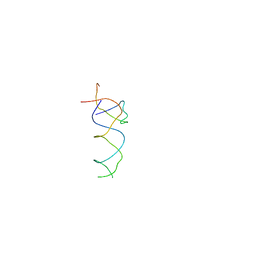 | | Components of the control 70S ribosome to provide reference for the RRF binding site | | Descriptor: | 19-mer fragment of the 23S rRNA, 42-mer fragment of double helix from 16S rRNA, dodecamer fragment of double helix from 23S rRNA | | Authors: | Agrawal, R.K, Sharma, M.R, Kiel, M.C, Hirokawa, G, Booth, T.M, Spahn, C.M, Grassucci, R.A, Kaji, A, Frank, J. | | Deposit date: | 2004-04-16 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2DT1
 
 | | Crystal Structure Of The Complex Of Goat Signalling Protein With Tetrasaccharide At 2.09 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kumar, J, Ethayathulla, A.S, Srivastava, D.B, Singh, N, Sharma, S, Bhushan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2006-07-09 | | Release date: | 2006-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Carbohydrate-binding properties of goat secretory glycoprotein (SPG-40) and its functional implications: structures of the native glycoprotein and its four complexes with chitin-like oligosaccharides
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2EWL
 
 | |
2DSV
 
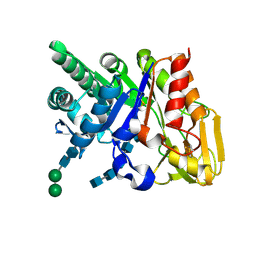 | | Interactions of protective signalling factor with chitin-like polysaccharide: Crystal structure of the complex between signalling protein from sheep (SPS-40) and a hexasaccharide at 2.5A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, alpha-D-mannopyranose-(1-4)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Srivastava, D.B, Ethayathulla, A.S, Kumar, J, Singh, N, Sharma, S, Bhushan, A, Srinivasan, A, Singh, T.P. | | Deposit date: | 2006-07-07 | | Release date: | 2006-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Carbohydrate binding properties and carbohydrate induced conformational switch in sheep secretory glycoprotein (SPS-40): crystal structures of four complexes of SPS-40 with chitin-like oligosaccharides
J.Struct.Biol., 158, 2007
|
|
3VHX
 
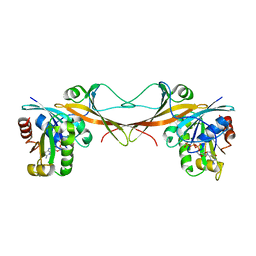 | | The crystal structure of Arf6-MKLP1 (Mitotic kinesin-like protein 1) complex | | Descriptor: | ADP-ribosylation factor 6, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Makyio, H, Takei, T, Ohgi, H, Takahashi, S, Takatsu, H, Ueda, T, Kanaho, Y, Xie, Y, Shin, H.W, Kamikubo, H, Kataoka, M, Kawasaki, M, Kato, R, Wakatsuki, S, Nakayama, K. | | Deposit date: | 2011-09-12 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for Arf6-MKLP1 complex formation on the Flemming body responsible for cytokinesis
Embo J., 31, 2012
|
|
2M9U
 
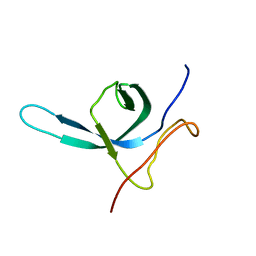 | | Solution NMR structure of the C-terminal domain (CTD) of Moloney murine leukemia virus integrase, Northeast Structural Genomics Target OR41A | | Descriptor: | Integrase p46 | | Authors: | Aiyer, S, Rossi, P, Schneider, W.M, Chander, A, Roth, M.J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-19 | | Release date: | 2013-12-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altering murine leukemia virus integration through disruption of the integrase and BET protein family interaction.
Nucleic Acids Res., 42, 2014
|
|
8UBY
 
 | | Choline-bound FLVCR1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, CHOLINE ION, Heme transporter FLVCR1 | | Authors: | Hite, R.K, Son, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1.
Nature, 629, 2024
|
|
8UBZ
 
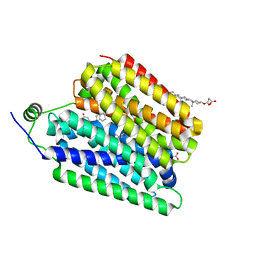 | | Choline-bound FLVCR1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, CHOLINE ION, Heme transporter FLVCR1 | | Authors: | Hite, R.K, Son, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1.
Nature, 629, 2024
|
|
8UBX
 
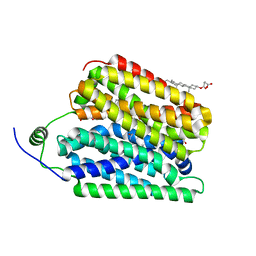 | | Ethanolamine-bound FLVCR1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, ETHANOLAMINE, Heme transporter FLVCR1 | | Authors: | Hite, R.K, Son, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1.
Nature, 629, 2024
|
|
8UC0
 
 | | Endogenous ligand bound FLVCR1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Heme transporter FLVCR1 | | Authors: | Hite, R.K, Son, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1.
Nature, 629, 2024
|
|
8UBW
 
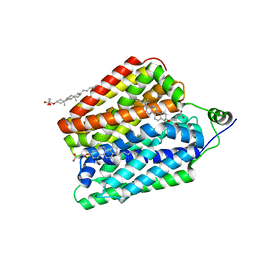 | | Choline-bound FLVCR1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, CHOLINE ION, Heme transporter FLVCR1 | | Authors: | Hite, R.K, Son, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1.
Nature, 629, 2024
|
|
8TWG
 
 | |
8TWF
 
 | |
