4INA
 
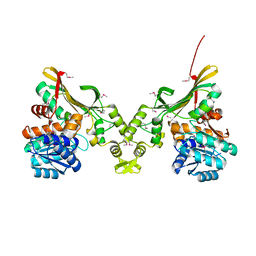 | | Crystal Structure of the Q7MSS8_WOLSU protein from Wolinella succinogenes. Northeast Structural Genomics Consortium Target WsR35 | | Descriptor: | saccharopine dehydrogenase | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-01-04 | | Release date: | 2013-01-16 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (2.488 Å) | | Cite: | Crystal Structure of the Q7MSS8_WOLSU protein from Wolinella succinogenes.
To be Published
|
|
6FCJ
 
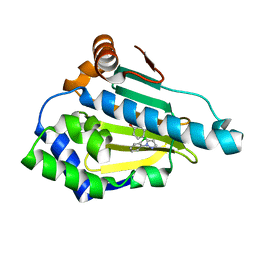 | |
2J1U
 
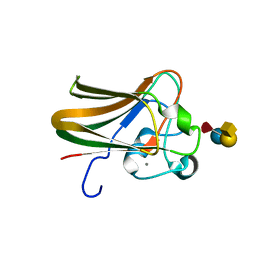 | |
6FGT
 
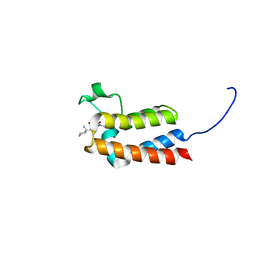 | | Crystal Structure of BAZ2B bromodomain in complex with 1-methylpyridinone compound 3 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, ~{N}-[(1-azanylcyclohexyl)methyl]-1-methyl-6-oxidanylidene-pyridine-3-carboxamide | | Authors: | Dalle Vedove, A, Spiliotopoulos, D, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
6FHP
 
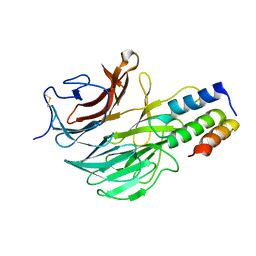 | | DAIP in complex with a C-terminal fragment of thermolysin | | Descriptor: | Dispase autolysis-inducing protein, Thermolysin | | Authors: | Schmelz, S, Fiebig, D, Fuchsbauer, H.L, Blankenfeldt, W, Scrima, A. | | Deposit date: | 2018-01-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Destructive twisting of neutral metalloproteases: the catalysis mechanism of the Dispase autolysis-inducing protein from Streptomyces mobaraensis DSM 40487.
FEBS J., 285, 2018
|
|
6F8E
 
 | | PH domain from TgAPH | | Descriptor: | Pleckstrin homology domain | | Authors: | Darvill, N, Liu, B, Matthews, S, Soldati-Favre, D, Rouse, S, Benjamin, S, Blake, T, Dubois, D.J, Hammoudi, P.M, Pino, P. | | Deposit date: | 2017-12-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Phosphatidic Acid Sensing by APH in Apicomplexan Parasites.
Structure, 26, 2018
|
|
2J44
 
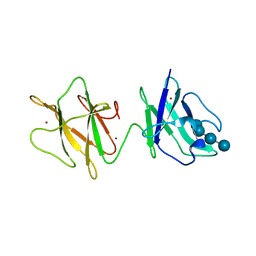 | | Alpha-glucan binding by a streptococcal virulence factor | | Descriptor: | ALKALINE AMYLOPULLULANASE, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Lammerts van Bueren, A, Higgins, M, Wang, D, Burke, R.D, Boraston, A.B. | | Deposit date: | 2006-08-24 | | Release date: | 2006-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and Structural Basis of Binding to Host Lung Glycogen by Streptococcal Virulence Factors.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2J4B
 
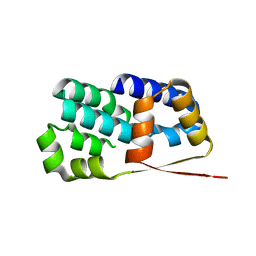 | | Crystal structure of Encephalitozoon cuniculi TAF5 N-terminal domain | | Descriptor: | TRANSCRIPTION INITIATION FACTOR TFIID SUBUNIT 72/90-100 KDA | | Authors: | Romier, C, James, N, Birck, C, Cavarelli, J, Vivares, C, Collart, M.A, Moras, D. | | Deposit date: | 2006-08-28 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure, Biochemical and Genetic Characterization of Yeast and E. Cuniculi Taf(II)5 N-Terminal Domain: Implications for TFIID Assembly.
J.Mol.Biol., 368, 2007
|
|
2J4I
 
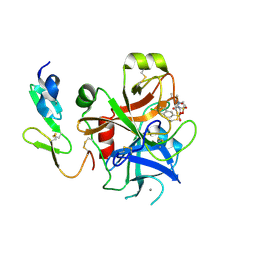 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 1-PYRROLIDINEACETAMIDE, 3-[[(6-CHLORO-2-NAPHTHALENYL)SULFONYL]AMINO]-ALPHA-METHYL-N-(1-METHYLETHYL)-N-[2-[(METHYLSULFONYL)AMINO]ETHYL]-2-OXO-, (ALPHAS,3S)-, ... | | Authors: | Young, R.J, Campbell, M, Borthwick, A.D, Brown, D, Chan, C, Convery, M.A, Crowe, M.C, Dayal, S, Diallo, H, Kelly, H.A, Paul King, N, Kleanthous, S, Kurtis, C.L, Mason, A.M, Mordaunt, J.E, Patel, C, Pateman, A.J, Senger, S, Shah, G.P, Smith, P.W, Watson, N.S, Weston, H.E, Zhou, P. | | Deposit date: | 2006-08-31 | | Release date: | 2006-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure- and Property-Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Acyclic Alanyl Amides as P4 Motifs.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
6FGE
 
 | | Crystal structure of human ZUFSP/ZUP1 in complex with ubiquitin | | Descriptor: | ACETATE ION, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kwasna, D, Abdul Rehman, S.A, Kulathu, Y. | | Deposit date: | 2018-01-10 | | Release date: | 2018-04-04 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery and Characterization of ZUFSP/ZUP1, a Distinct Deubiquitinase Class Important for Genome Stability.
Mol. Cell, 70, 2018
|
|
6FGV
 
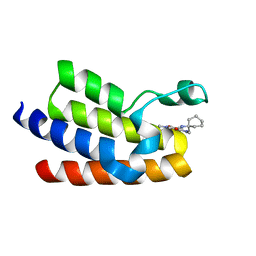 | | Crystal Structure of BAZ2A bromodomain in complex with 1-methylpyridinone compound 3 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, ~{N}-[(1-azanylcyclohexyl)methyl]-1-methyl-6-oxidanylidene-pyridine-3-carboxamide | | Authors: | Dalle Vedove, A, Spiliotopoulos, D, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
6EYT
 
 | |
2J1M
 
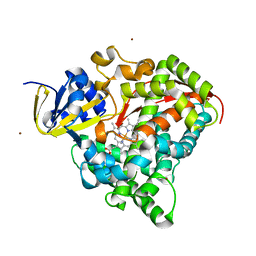 | | P450 BM3 Heme domain in complex with DMSO | | Descriptor: | CYTOCHROME P450 102, DIMETHYL SULFOXIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kuper, J, Tuck-Seng, W, Roccatano, D, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2006-08-14 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Understanding a Mechanism of Organic Cosolvent Inactivation in Heme Monooxygenase P450 Bm-3.
J.Am.Chem.Soc., 129, 2007
|
|
2IUC
 
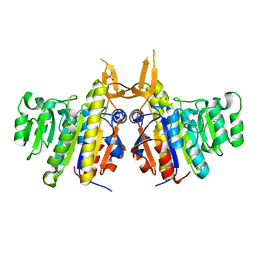 | | Structure of alkaline phosphatase from the Antarctic bacterium TAB5 | | Descriptor: | ALKALINE PHOSPHATASE, CACODYLATE ION, MAGNESIUM ION, ... | | Authors: | Wang, E, Koutsioulis, D, Leiros, H.K.S, Andersen, O.A, Bouriotis, V, Hough, E, Heikinheimo, P. | | Deposit date: | 2006-06-01 | | Release date: | 2006-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Alkaline Phosphatase from the Antarctic Bacterium Tab5.
J.Mol.Biol., 366, 2007
|
|
2J2Z
 
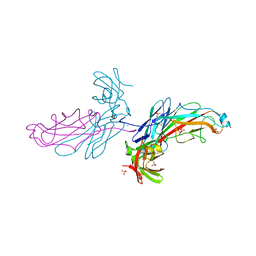 | | X-Ray Structure of the Chaperone PapD in complex with the Pilus terminator subunit PapH at 2.3 Angstrom resolution | | Descriptor: | CHAPERONE PROTEIN PAPD, COBALT (II) ION, PAP FIMBRIAL MINOR PILIN PROTEIN, ... | | Authors: | Verger, D, Miller, E, Remaut, H, Waksman, G, Hultgren, S. | | Deposit date: | 2006-08-17 | | Release date: | 2006-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Mechanism of P Pilus Termination in Uropathogenic Escherichia Coli.
Embo Rep., 7, 2006
|
|
2J0M
 
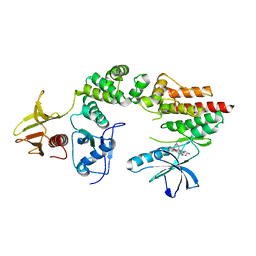 | | Crystal structure a two-chain complex between the FERM and kinase domains of focal adhesion kinase. | | Descriptor: | 1,2,3,4-TETRAHYDROGEN-STAUROSPORINE, FOCAL ADHESION KINASE 1 | | Authors: | Lietha, D, Cai, X, Li, Y, Schaller, M.D, Eck, M.J. | | Deposit date: | 2006-08-03 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Autoinhibition of Focal Adhesion Kinase.
Cell(Cambridge,Mass.), 129, 2007
|
|
6F8Y
 
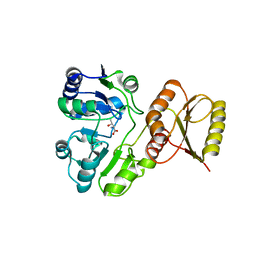 | | Crystal structure of P. abyssi Sua5 complexed with L-threonine | | Descriptor: | THREONINE, Threonylcarbamoyl-AMP synthase | | Authors: | Pichard-Kostuch, A, Zhang, W, Liger, D, Daugeron, M.C, Letoquart, J, Li de la Sierra-Gallay, I, Forterre, P, Collinet, B, van Tilbeurgh, H, Basta, T. | | Deposit date: | 2017-12-13 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure-function analysis of Sua5 protein reveals novel functional motifs required for the biosynthesis of the universal t6A tRNA modification.
RNA, 24, 2018
|
|
2J5C
 
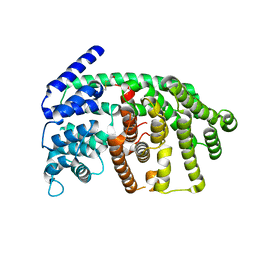 | | Rational conversion of substrate and product specificity in a monoterpene synthase. Structural insights into the molecular basis of rapid evolution. | | Descriptor: | 1,8-CINEOLE SYNTHASE, BETA-MERCAPTOETHANOL | | Authors: | Kampranis, S.C, Ioannidis, D, Purvis, A, Mahrez, W, Ninga, E, Katerelos, N.A, Anssour, S, Dunwell, J.M, Makris, A.M, Goodenough, P.W, Johnson, C.B. | | Deposit date: | 2006-09-14 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational Conversion of Substrate and Product Specificity in a Salvia Monoterpene Synthase: Structural Insights Into the Evolution of Terpene Synthase Function.
Plant Cell, 19, 2007
|
|
6EZG
 
 | | Torpedo californica AChE in complex with indolic multi-target directed ligand | | Descriptor: | 1-(7-chloranyl-4-methoxy-1~{H}-indol-5-yl)-3-[1-(phenylmethyl)piperidin-4-yl]propan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Santoni, G, Lalut, J, Karila, D, Lecoutey, C, Davis, A, Nachon, F, Sillman, I, Sussman, J, Weik, M, Maurice, T, Dallemagne, P, Rochais, C. | | Deposit date: | 2017-11-15 | | Release date: | 2018-11-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel multitarget-directed ligands targeting acetylcholinesterase and sigma1receptors as lead compounds for treatment of Alzheimer's disease: Synthesis, evaluation, and structural characterization of their complexes with acetylcholinesterase.
Eur J Med Chem, 162, 2018
|
|
2J0K
 
 | | Crystal structure of a fragment of focal adhesion kinase containing the FERM and kinase domains. | | Descriptor: | 1,2,3,4-TETRAHYDROGEN-STAUROSPORINE, FOCAL ADHESION KINASE 1 | | Authors: | Lietha, D, Cai, X, Li, Y, Schaller, M.D, Eck, M.J. | | Deposit date: | 2006-08-03 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for the Autoinhibition of Focal Adhesion Kinase.
Cell(Cambridge,Mass.), 129, 2007
|
|
4EAI
 
 | | Co-crystal structure of an AMPK core with AMP | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Chen, L, Wang, J, Zhang, Y.-Y, Yan, S.F, Neumann, D, Schlattner, U, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | AMP-activated protein kinase undergoes nucleotide-dependent conformational changes
Nat.Struct.Mol.Biol., 19, 2012
|
|
6FCZ
 
 | | Model of gC1q-Fc complex based on 7A EM map | | Descriptor: | Complement C1q subcomponent subunit A, Complement C1q subcomponent subunit B, Complement C1q subcomponent subunit C, ... | | Authors: | Ugurlar, D, Howes, S.C, de Kreuk, B.J.K, de Jong, R.N, Beurskens, F.J, Koster, A.J, Parren, P.W.H.I, Sharp, T.H, Gros, P, Koning, R.I. | | Deposit date: | 2017-12-21 | | Release date: | 2018-02-28 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structures of C1-IgG1 provide insights into how danger pattern recognition activates complement.
Science, 359, 2018
|
|
4E4E
 
 | |
2J0J
 
 | | Crystal structure of a fragment of focal adhesion kinase containing the FERM and kinase domains. | | Descriptor: | 1,2,3,4-TETRAHYDROGEN-STAUROSPORINE, FOCAL ADHESION KINASE 1 | | Authors: | Lietha, D, Cai, X, Li, Y, Schaller, M.D, Eck, M.J. | | Deposit date: | 2006-08-03 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Autoinhibition of Focal Adhesion Kinase
Cell(Cambridge,Mass.), 129, 2007
|
|
5NJ5
 
 | | E. coli Microcin-processing metalloprotease TldD/E with phosphate bound | | Descriptor: | 1,2-ETHANEDIOL, Metalloprotease PmbA, Metalloprotease TldD, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
