5EHR
 
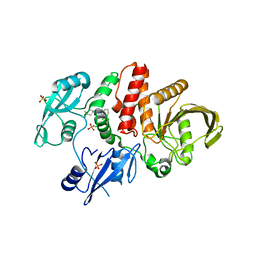 | |
6SEQ
 
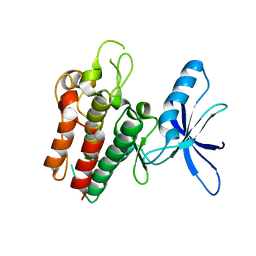 | | Lemur tyrosine kinase 3 (LMTK3) | | Descriptor: | Serine/threonine-protein kinase LMTK3 | | Authors: | Roe, S.M, Owen, R. | | Deposit date: | 2019-07-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure-function relationship of oncogenic LMTK3.
Sci Adv, 6, 2020
|
|
3QME
 
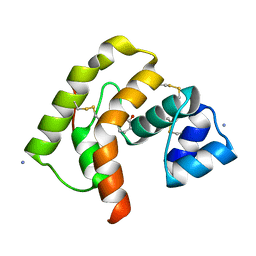 | |
4JAM
 
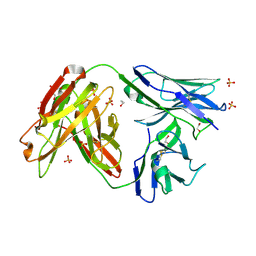 | | Crystal structure of broadly neutralizing anti-hiv-1 antibody ch103 | | Descriptor: | 1,2-ETHANEDIOL, ANTIGEN BINDING FRAGMENT OF HEAVY CHAIN of CH103, ANTIGEN BINDING FRAGMENT OF LIGHT CHAIN of CH103, ... | | Authors: | Zhou, T, Moquin, S, Zheng, A, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus.
Nature, 496, 2013
|
|
1NZY
 
 | |
8KHU
 
 | | Hepatitis B virus core protein Y132A mutant in complex with THPP derivatives 48 | | Descriptor: | (6~{S},7~{R})-6,7-dimethyl-3-(2-oxidanylidenepyrrolidin-1-yl)-~{N}-[3,4,5-tris(fluoranyl)phenyl]-6,7-dihydro-4~{H}-pyrazolo[1,5-a]pyrazine-5-carboxamide, Capsid protein, GLYCEROL, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2023-08-22 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4,5,6,7-Tetrahydropyrazolo[1.5-a]pyrizine Derivatives as Core Protein Allosteric Modulators (CpAMs) for the Inhibition of Hepatitis B Virus.
J.Med.Chem., 66, 2023
|
|
4K6H
 
 | | Crystal structure of CALB mutant L278M from Candida antarctica | | Descriptor: | 1,2-ETHANEDIOL, Lipase B | | Authors: | An, J, Xie, Y, Feng, Y, Wu, G. | | Deposit date: | 2013-04-15 | | Release date: | 2014-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhanced enzyme kinetic stability by increasing rigidity within the active site.
J.Biol.Chem., 289, 2014
|
|
5FN2
 
 | | Cryo-EM structure of gamma secretase in complex with a drug DAPT | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
5FN3
 
 | | Cryo-EM structure of gamma secretase in class 1 of the apo- state ensemble | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
4K5Q
 
 | |
4K6K
 
 | | Crystal structure of CALB mutant D223G from Candida antarctica | | Descriptor: | 1,2-ETHANEDIOL, Lipase B | | Authors: | An, J, Xie, Y, Feng, Y, Wu, G. | | Deposit date: | 2013-04-16 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhanced enzyme kinetic stability by increasing rigidity within the active site.
J.Biol.Chem., 289, 2014
|
|
4K6G
 
 | | Crystal structure of CALB from Candida antarctica | | Descriptor: | 1,2-ETHANEDIOL, Lipase B | | Authors: | An, J, Xie, Y, Feng, Y, Wu, G. | | Deposit date: | 2013-04-15 | | Release date: | 2014-01-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhanced enzyme kinetic stability by increasing rigidity within the active site.
J.Biol.Chem., 289, 2014
|
|
4LVG
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | (1S,2S)-N-[4-(phenylsulfonyl)phenyl]-2-(pyridin-3-yl)cyclopropanecarboxamide, 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5WTW
 
 | |
5WRE
 
 | | Hepatitis B virus core protein Y132A mutant in complex with heteroaryldihydropyrimidine (HAP_R01) | | Descriptor: | (2S)-1-[[(4R)-4-(2-chloranyl-4-fluoranyl-phenyl)-5-methoxycarbonyl-2-(1,3-thiazol-2-yl)-1,4-dihydropyrimidin-6-yl]methyl]-4,4-bis(fluoranyl)pyrrolidine-2-carboxylic acid, CHLORIDE ION, Core protein, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2016-12-01 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | Heteroaryldihydropyrimidine (HAP) and Sulfamoylbenzamide (SBA) Inhibit Hepatitis B Virus Replication by Different Molecular Mechanisms.
Sci Rep, 7, 2017
|
|
6O6Y
 
 | | Crystal structure of Csm6 in complex with cyclic-tetraadenylates (cA4) by cocrystallization of Csm6 and cA4 | | Descriptor: | 2',3'- cyclic AMP, 3'-O-[(R)-{[(2S,3aS,4S,6S,6aS)-6-(6-amino-9H-purin-9-yl)-2-hydroxy-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]methoxy}(hydroxy)phosphoryl]adenosine, Csm6 | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | CRISPR-Cas III-A Csm6 CARF Domain Is a Ring Nuclease Triggering Stepwise cA4Cleavage with ApA>p Formation Terminating RNase Activity.
Mol.Cell, 75, 2019
|
|
6O6S
 
 | | Crystal structure of Apo Csm6 | | Descriptor: | Csm6 | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | CRISPR-Cas III-A Csm6 CARF Domain Is a Ring Nuclease Triggering Stepwise cA4Cleavage with ApA>p Formation Terminating RNase Activity.
Mol.Cell, 75, 2019
|
|
6OV0
 
 | |
4QS4
 
 | |
6O6X
 
 | |
6O6V
 
 | |
6O70
 
 | | Crystal structure of Csm6 H132A mutant in complex with cA4 by cocrystallization of cA4 and Csm6 H132A mutant | | Descriptor: | 2',3'- cyclic AMP, 3'-O-[(R)-{[(2S,3aS,4S,6S,6aS)-6-(6-amino-9H-purin-9-yl)-2-hydroxy-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]methoxy}(hydroxy)phosphoryl]adenosine, Csm6 | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRISPR-Cas III-A Csm6 CARF Domain Is a Ring Nuclease Triggering Stepwise cA4Cleavage with ApA>p Formation Terminating RNase Activity.
Mol.Cell, 75, 2019
|
|
6O6Z
 
 | | Crystal structure of Csm6 H381A in complex with cA4 by cocrystallization of cA4 and Csm6 | | Descriptor: | 3'-O-[(R)-{[(2S,3aS,4S,6S,6aS)-6-(6-amino-9H-purin-9-yl)-2-hydroxy-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]methoxy}(hydroxy)phosphoryl]adenosine, Csm6 | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRISPR-Cas III-A Csm6 CARF Domain Is a Ring Nuclease Triggering Stepwise cA4Cleavage with ApA>p Formation Terminating RNase Activity.
Mol.Cell, 75, 2019
|
|
2VAY
 
 | | Calmodulin complexed with CaV1.1 IQ peptide | | Descriptor: | CALCIUM ION, CALMODULIN, CHLORIDE ION, ... | | Authors: | Halling, D.B, Black, D.J, Pedersen, S.E, Hamilton, S.L. | | Deposit date: | 2007-09-05 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Determinants in Cav1 Channels that Regulate the Ca2+ Sensitivity of Bound Calmodulin.
J.Biol.Chem., 284, 2009
|
|
3G43
 
 | |
