2PQT
 
 | | Human N-acetyltransferase 1 | | Descriptor: | Arylamine N-acetyltransferase 1, CHLORIDE ION, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Wu, H, Dombrovski, L, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Grant, D.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-02 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis of Substrate-binding Specificity of Human Arylamine N-Acetyltransferases.
J.Biol.Chem., 282, 2007
|
|
2PQ8
 
 | | MYST histone acetyltransferase 1 | | Descriptor: | COENZYME A, Probable histone acetyltransferase MYST1, UNKNOWN ATOM OR ION, ... | | Authors: | Tempel, W, Wu, H, Dombrovski, L, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | MYST histone acetyltransferase 1.
To be Published
|
|
2PFR
 
 | | Human N-acetyltransferase 2 | | Descriptor: | Arylamine N-acetyltransferase 2, COENZYME A, SULFATE ION, ... | | Authors: | Tempel, W, Wu, H, Dombrovski, L, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Grant, D.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-05 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The Crystal Structure of Human N-acetyltransferase 2 in complex with CoA
to be published
|
|
5WOC
 
 | |
6XKK
 
 | | Cryo-EM structure of the NLRP1-CARD filament | | Descriptor: | NACHT, LRR and PYD domains-containing protein 1 | | Authors: | Hollingsworth, L.R, David, L, Li, Y, Sharif, H, Fontana, P, Fu, T, Wu, H. | | Deposit date: | 2020-06-26 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Mechanism of filament formation in UPA-promoted CARD8 and NLRP1 inflammasomes.
Nat Commun, 12, 2021
|
|
6XKJ
 
 | | Cryo-EM structure of CARD8-CARD filament | | Descriptor: | Caspase recruitment domain-containing protein 8 | | Authors: | Hollingsworth, L.R, David, L, Li, Y, Sharif, H, Fontana, P, Fu, T, Wu, H. | | Deposit date: | 2020-06-26 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Mechanism of filament formation in UPA-promoted CARD8 and NLRP1 inflammasomes.
Nat Commun, 12, 2021
|
|
3DB5
 
 | | Crystal structure of methyltransferase domain of human PR domain-containing protein 4 | | Descriptor: | PR domain zinc finger protein 4 | | Authors: | Amaya, M.F, Zeng, H, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Plotnikov, A.N, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-30 | | Release date: | 2008-08-12 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of methyltransferase domain of human PR domain-containing protein 4.
To be Published
|
|
2FXF
 
 | | Human spermidine/spermine N1-acetyltransferase | | Descriptor: | ACETATE ION, ACETYL COENZYME *A, CALCIUM ION, ... | | Authors: | Min, J.R, Wu, H, Zeng, H, Loppnau, P, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-02-05 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human spermidine/spermine
N1-acetyltransferase
To be Published
|
|
6DLP
 
 | | Crystal structure of LRRK2 WD40 domain dimer | | Descriptor: | Leucine-rich repeat serine/threonine-protein kinase 2, PLATINUM (II) ION | | Authors: | Zhang, P, Ru, H, Wang, L, Wu, H. | | Deposit date: | 2018-06-02 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structure of the WD40 domain dimer of LRRK2.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4PHR
 
 | | Domain of unknown function 1792 (DUF1792) with manganese | | Descriptor: | ACETATE ION, MANGANESE (II) ION, Putative glycosyltransferase (GalT1), ... | | Authors: | Zhang, H, Wu, H. | | Deposit date: | 2014-05-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | The highly conserved domain of unknown function 1792 has a distinct glycosyltransferase fold.
Nat Commun, 5, 2014
|
|
4PFX
 
 | |
4PHS
 
 | |
5WCF
 
 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | (5R)-4-(5-bromofuran-2-carbonyl)-5-(4-fluorophenyl)-7-methyl-1,3,4,5-tetrahydro-2H-1,4-benzodiazepin-2-one, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Dong, A, Zeng, H, Hutch, A, Seitova, A, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Crystal Structure of Human HMT1 hnRNP methyltransferase-like protein 6 in complex with MTLLE1441
to be published
|
|
2FW2
 
 | | Catalytic domain of CDY | | Descriptor: | Testis-specific chromodomain protein Y 2 | | Authors: | Min, J.R, Antoshenko, T, Wu, H, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-31 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of catalytic domain of Human Testis-specific chromodomain protein Y.
To be Published
|
|
3M0A
 
 | | Crystal structure of TRAF2:cIAP2 complex | | Descriptor: | Baculoviral IAP repeat-containing protein 3, TNF receptor-associated factor 2, ZINC ION | | Authors: | Kabaleeswaran, V, Wu, H. | | Deposit date: | 2010-03-02 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structures of the TRAF2: cIAP2 and the TRAF1: TRAF2: cIAP2 complexes: affinity, specificity, and regulation.
Mol.Cell, 38, 2010
|
|
3MEK
 
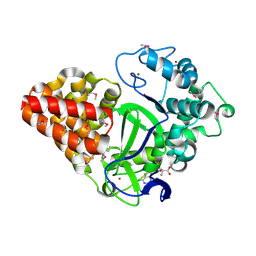 | | Crystal Structure of Human Histone-Lysine N-methyltransferase SMYD3 in Complex with S-adenosyl-L-methionine | | Descriptor: | S-ADENOSYLMETHIONINE, SET and MYND domain-containing protein 3, ZINC ION | | Authors: | Lam, R, Dombrovski, L, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Histone-Lysine N-methyltransferase SMYD3 in Complex with S-adenosyl-L-methionine
To be Published
|
|
3MOP
 
 | | The ternary Death Domain complex of MyD88, IRAK4, and IRAK2 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, Interleukin-1 receptor-associated kinase-like 2, Myeloid differentiation primary response protein MyD88 | | Authors: | Lin, S.-C, Lo, Y.-C, Wu, H. | | Deposit date: | 2010-04-23 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling.
Nature, 465, 2010
|
|
6EGA
 
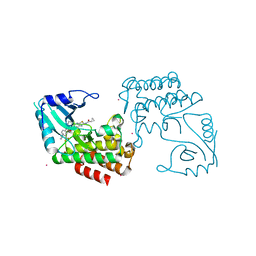 | | IRAK4 in complex with a type II inhibitor | | Descriptor: | 3-{2-[(cyclopropanecarbonyl)amino][1,3]thiazolo[5,4-b]pyridin-5-yl}-N-{4-[(piperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide, COBALT (II) ION, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Wang, L, Wu, H. | | Deposit date: | 2018-08-19 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.512 Å) | | Cite: | Conformational flexibility and inhibitor binding to unphosphorylated interleukin-1 receptor-associated kinase 4 (IRAK4).
J.Biol.Chem., 294, 2019
|
|
7SEK
 
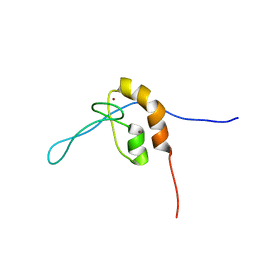 | | Solution structure of the zinc finger domain of murine MetAP1, complexed with ZNG N-terminal peptide | | Descriptor: | COBW domain-containing protein 1,Methionine aminopeptidase 1 fusion, ZINC ION | | Authors: | Edmonds, K.A, Jordan, M.R, Thalluri, K, Wu, H, Di Marchi, R, Giedroc, D.P. | | Deposit date: | 2021-09-30 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Zn-regulated GTPase metalloprotein activator 1 modulates vertebrate zinc homeostasis.
Cell, 185, 2022
|
|
4W6Q
 
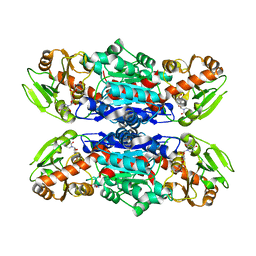 | | Glycosyltransferase C from Streptococcus agalactiae | | Descriptor: | SERINE, URIDINE-5'-DIPHOSPHATE, glucosyltransferase | | Authors: | Zhu, F, Zhang, H, Wu, H. | | Deposit date: | 2014-08-20 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A conserved domain is crucial for acceptor substrate binding in a family of glucosyltransferases.
J.Bacteriol., 197, 2015
|
|
4PQG
 
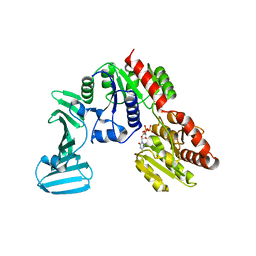 | | Crystal structure of the pneumococcal O-GlcNAc transferase GtfA in complex with UDP and GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosyltransferase Gtf1, URIDINE-5'-DIPHOSPHATE | | Authors: | Shi, W.W, Jiang, Y.L, Zhu, F, Yang, Y.H, Wu, H, Ren, Y.M, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Novel O-Linked N-Acetyl-d-glucosamine (O-GlcNAc) Transferase, GtfA, Reveals Insights into the Glycosylation of Pneumococcal Serine-rich Repeat Adhesins.
J.Biol.Chem., 289, 2014
|
|
5W0U
 
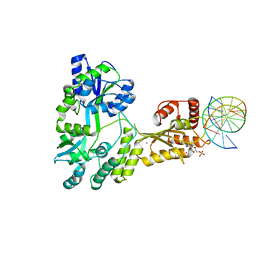 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with dCMP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
5W1C
 
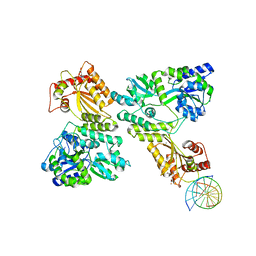 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
6L62
 
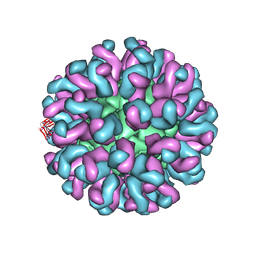 | | Neutralization mechanism of a monoclonal antibody targeting a porcine circovirus type 2 Cap protein conformational epitope | | Descriptor: | Capsid protein, Heavy chain of Fab fragment, Light chain of Fab fragment | | Authors: | Sun, Z, Huang, L, Xia, D, Wei, Y, Sun, E, Zhu, H, Bian, H, Wu, H, Feng, L, Wang, J, Liu, C. | | Deposit date: | 2019-10-25 | | Release date: | 2020-02-12 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Neutralization Mechanism of a Monoclonal Antibody Targeting a Porcine Circovirus Type 2 Cap Protein Conformational Epitope.
J.Virol., 94, 2020
|
|
6LM3
 
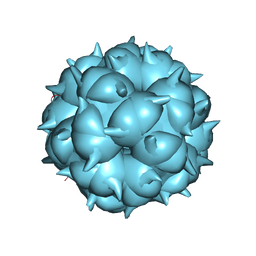 | | Neutralization mechanism of a monoclonal antibody targeting a porcine circovirus type 2 Cap protein conformational epitope | | Descriptor: | Capsid protein | | Authors: | Sun, Z, Huang, L, Xia, D, Wei, Y, Sun, E, Zhu, H, Bian, H, Wu, H, Feng, L, Wang, J, Liu, C. | | Deposit date: | 2019-12-24 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Neutralization Mechanism of a Monoclonal Antibody Targeting a Porcine Circovirus Type 2 Cap Protein Conformational Epitope.
J.Virol., 94, 2020
|
|
