6JB8
 
 | | Crystal structure of nanobody D3-L11 in complex with hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Caaveiro, J.M.M, Tamura, H, Akiba, H, Tsumoto, K. | | Deposit date: | 2019-01-25 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and thermodynamic basis for the recognition of the substrate-binding cleft on hen egg lysozyme by a single-domain antibody.
Sci Rep, 9, 2019
|
|
6JB5
 
 | | Crystal structure of nanobody D3-L11 mutant Y102A in complex with hen egg-white lysozyme (form II) | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Caaveiro, J.M.M, Tamura, H, Akiba, H, Tsumoto, K. | | Deposit date: | 2019-01-25 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and thermodynamic basis for the recognition of the substrate-binding cleft on hen egg lysozyme by a single-domain antibody.
Sci Rep, 9, 2019
|
|
6AFB
 
 | | DJ-1 C106S incubated with isatin | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, ISATIN, ... | | Authors: | Caaveiro, J.M.M, Tashiro, S, Tsumoto, K. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
ACS Chem. Biol., 13, 2018
|
|
6AFL
 
 | | DJ-1 with compound 15 | | Descriptor: | 5-fluoranyl-1-(2-phenylethyl)indole-2,3-dione, CHLORIDE ION, Protein/nucleic acid deglycase DJ-1 | | Authors: | Caaveiro, J.M.M, Tashiro, S, Tsumoto, K. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
ACS Chem. Biol., 13, 2018
|
|
8J81
 
 | | MDM2 bound with a peptoid | | Descriptor: | (2S)-2-[[(2S)-2-[(6-chloranyl-1H-indol-3-yl)methyl-[(2S)-2-[[(2S)-2-[ethanoyl-(phenylmethyl)amino]propanoyl]-methyl-amino]propanoyl]amino]propanoyl]-methyl-amino]-N-(3,3-dimethylbutyl)-N-[(2S)-1-oxidanylidene-1-piperazin-1-yl-propan-2-yl]propanamide, CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Yokomine, M, Fukuda, Y, Ago, H, Matsuura, H, Ueno, G, Nagatoishi, S, Yamamoto, M, Tsumoto, K, Jumpei, M, Sando, S. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A high-resolution structural characterization and physicochemical study of how a peptoid binds to an oncoprotein MDM2.
Chem Sci, 15, 2024
|
|
5CI9
 
 | | Crystal structure of human Tob in complex with inhibitor fragment 6 | | Descriptor: | 1-(propan-2-yl)-1H-benzimidazole-5-carboxylic acid, Protein Tob1, SODIUM ION | | Authors: | Bai, Y, Tashiro, S, Nagatoishi, S, Suzuki, T, Tsumoto, K, Bartlam, M, Yamamoto, T. | | Deposit date: | 2015-07-11 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for inhibition of the Tob-CNOT7 interaction by a fragment screening approach
Protein Cell, 6, 2015
|
|
5CI8
 
 | | Crystal structure of human Tob in complex with inhibitor fragment 1 | | Descriptor: | Protein Tob1, pyrrolo[1,2-a]quinoxalin-4(5H)-one | | Authors: | Bai, Y, Tashiro, S, Nagatoishi, S, Suzuki, T, Tsumoto, K, Bartlam, M, Yamamoto, T. | | Deposit date: | 2015-07-11 | | Release date: | 2015-11-18 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Structural basis for inhibition of the Tob-CNOT7 interaction by a fragment screening approach
Protein Cell, 6, 2015
|
|
4YRD
 
 | | Crystal structure of CapF with inhibitor 3-isopropenyl-tropolone | | Descriptor: | 2-hydroxy-3-(prop-1-en-2-yl)cyclohepta-2,4,6-trien-1-one, Capsular polysaccharide synthesis enzyme Cap5F, ZINC ION | | Authors: | Nakano, K, Chigira, T, Miyafusa, T, Nagatoishi, S, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2015-03-15 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Discovery and characterization of natural tropolones as inhibitors of the antibacterial target CapF from Staphylococcus aureus.
Sci Rep, 5, 2015
|
|
5AZ2
 
 | | Crystal structure of the Fab fragment of 9E5, a murine monoclonal antibody specific for human epiregulin | | Descriptor: | anti-human epiregulin antibody 9E5 Fab heavy chain, anti-human epiregulin antibody 9E5 Fab light chain | | Authors: | Kado, Y, Mizohata, E, Nagatoishi, S, Iijima, M, Shinoda, K, Miyafusa, T, Nakayama, T, Yoshizumi, T, Sugiyama, A, Kawamura, T, Lee, Y.H, Matsumura, H, Doi, H, Fujitani, H, Kodama, T, Shibasaki, Y, Tsumoto, K, Inoue, T. | | Deposit date: | 2015-09-16 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
J.Biol.Chem., 291, 2016
|
|
5BTQ
 
 | | Crystal structure of human heme oxygenase 1 H25R with biliverdin bound | | Descriptor: | BILIVERDINE IX ALPHA, Heme oxygenase 1, SULFATE ION | | Authors: | Caaveiro, J.M.M, Morante, K, Sigala, P, Tsumoto, K. | | Deposit date: | 2015-06-03 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | In-Cell Enzymology To Probe His-Heme Ligation in Heme Oxygenase Catalysis
Biochemistry, 55, 2016
|
|
1UA6
 
 | | Crystal structure of HYHEL-10 FV MUTANT SFSF complexed with HEN EGG WHITE LYSOZYME complex | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Kumagai, I, Nishimiya, Y, Kondo, H, Tsumoto, K. | | Deposit date: | 2003-02-28 | | Release date: | 2004-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural consequences of target epitope-directed functional alteration of an antibody. The case of anti-hen lysozyme antibody, HyHEL-10
J.Biol.Chem., 278, 2003
|
|
1UD9
 
 | | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) Homolog From Sulfolobus tokodaii | | Descriptor: | DNA polymerase sliding clamp A, ZINC ION | | Authors: | Tanabe, E, Yasutake, Y, Tanaka, Y, Yao, M, Tsumoto, K, Kumagai, I, Tanaka, I. | | Deposit date: | 2003-04-28 | | Release date: | 2004-06-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) Homolog From Sulfolobus tokodaii
To be published
|
|
5Y78
 
 | | Crystal structure of the triose-phosphate/phosphate translocator in complex with inorganic phosphate | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PHOSPHATE ION, Putative hexose phosphate translocator | | Authors: | Lee, Y, Nishizawa, T, Takemoto, M, Kumazaki, K, Yamashita, K, Hirata, K, Minoda, A, Nagatoishi, S, Tsumoto, K, Ishitani, R, Nureki, O. | | Deposit date: | 2017-08-16 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the triose-phosphate/phosphate translocator reveals the basis of substrate specificity
Nat Plants, 3, 2017
|
|
5Y79
 
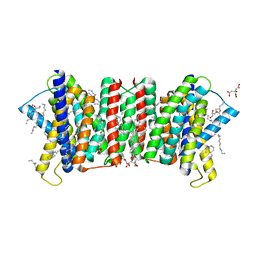 | | Crystal structure of the triose-phosphate/phosphate translocator in complex with 3-phosphoglycerate | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-PHOSPHOGLYCERIC ACID, CITRATE ANION, ... | | Authors: | Lee, Y, Nishizawa, T, Takemoto, M, Kumazaki, K, Yamashita, K, Hirata, K, Minoda, A, Nagatoishi, S, Tsumoto, K, Ishitani, R, Nureki, O. | | Deposit date: | 2017-08-16 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the triose-phosphate/phosphate translocator reveals the basis of substrate specificity
Nat Plants, 3, 2017
|
|
1UMJ
 
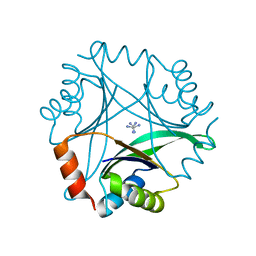 | | Crystal structure of Pyrococcus horikoshii CutA in the presence of 3M guanidine hydrochloride | | Descriptor: | GUANIDINE, periplasmic divalent cation tolerance protein CutA | | Authors: | Tanaka, Y, Tsumoto, K, Yasutake, Y, Sakai, N, Yao, M, Tanaka, I, Kumagai, I. | | Deposit date: | 2003-10-02 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural evidence for guanidine-protein side chain interactions: crystal structure of CutA from Pyrococcus horikoshii in 3M guanidine hydrochloride
Biochem.Biophys.Res.Commun., 323, 2004
|
|
1VBF
 
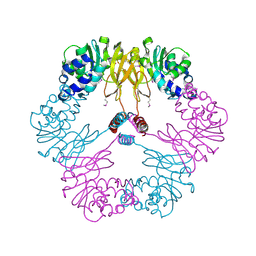 | | Crystal structure of protein L-isoaspartate O-methyltransferase homologue from Sulfolobus tokodaii | | Descriptor: | 231aa long hypothetical protein-L-isoaspartate O-methyltransferase | | Authors: | Tanaka, Y, Tsumoto, K, Yasutake, Y, Umetsu, M, Yao, M, Tanaka, I, Fukada, H, Kumagai, I. | | Deposit date: | 2004-02-25 | | Release date: | 2004-08-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | How Oligomerization Contributes to the Thermostability of an Archaeon Protein: PROTEIN L-ISOASPARTYL-O-METHYLTRANSFERASE FROM SULFOLOBUS TOKODAII
J.Biol.Chem., 279, 2004
|
|
1UAC
 
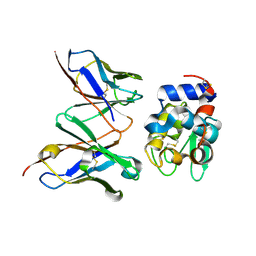 | | Crystal Structure of HYHEL-10 FV MUTANT SFSF Complexed with TURKEY WHITE LYSOZYME | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Kumagai, I, Nishimiya, Y, Kondo, H, Tsumoto, K. | | Deposit date: | 2003-03-08 | | Release date: | 2004-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural consequences of target epitope-directed functional alteration of an antibody. The case of anti-hen lysozyme antibody, HyHEL-10
J.BIOL.CHEM., 278, 2003
|
|
1UKU
 
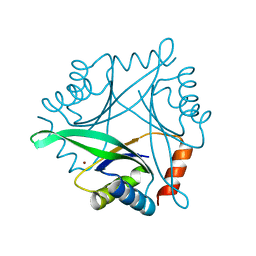 | | Crystal Structure of Pyrococcus horikoshii CutA1 Complexed with Cu2+ | | Descriptor: | COPPER (II) ION, periplasmic divalent cation tolerance protein CutA | | Authors: | Tanaka, Y, Yasutake, Y, Yao, M, Sakai, N, Tanaka, I, Tsumoto, K, Kumagai, I. | | Deposit date: | 2003-09-01 | | Release date: | 2004-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural implications for heavy metal-induced reversible assembly and aggregation of a protein: the case of Pyrococcus horikoshii CutA.
Febs Lett., 556, 2004
|
|
1VE0
 
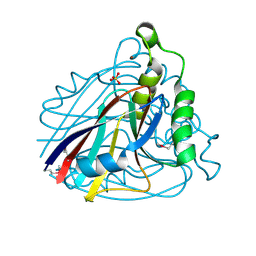 | | Crystal structure of uncharacterized protein ST2072 from Sulfolobus tokodaii | | Descriptor: | SULFATE ION, ZINC ION, hypothetical protein (ST2072) | | Authors: | Tanabe, E, Horiike, Y, Tsumoto, K, Yasutake, Y, Yao, M, Tanaka, I, Kumagai, I. | | Deposit date: | 2004-03-26 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the uncharacterized protein ST2072 from Sulfolobus tokodaii
To be Published
|
|
7CME
 
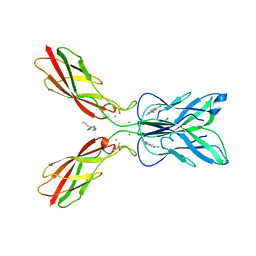 | | Crystal structure of human P-cadherin MEC12 (X dimer) in complex with 2-(5-chloro-2-methyl-1H-indol-3-yl)ethan-1-amine (inhibitor) | | Descriptor: | 2-(5-chloro-2-methyl-1H-indol-3-yl)ethan-1-amine, CALCIUM ION, Cadherin-3, ... | | Authors: | Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Regulation of cadherin dimerization by chemical fragments as a trigger to inhibit cell adhesion
Commun Biol, 4, 2021
|
|
7CMF
 
 | | Crystal structure of human P-cadherin REC12 (monomer) in complex with 2-(5-chloro-2-methyl-1H-indol-3-yl)ethan-1-amine (inhibitor) | | Descriptor: | 2-(5-chloro-2-methyl-1H-indol-3-yl)ethan-1-amine, CALCIUM ION, Cadherin-3 | | Authors: | Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regulation of cadherin dimerization by chemical fragments as a trigger to inhibit cell adhesion
Commun Biol, 4, 2021
|
|
3ST7
 
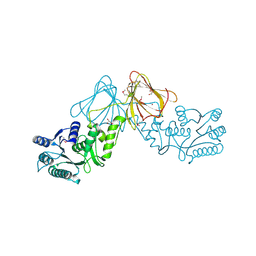 | | Crystal Structure of capsular polysaccharide assembling protein CapF from staphylococcus aureus | | Descriptor: | Capsular polysaccharide synthesis enzyme Cap5F, GLYCEROL, ZINC ION | | Authors: | Miyafusa, T, Tanaka, Y, Kuroda, M, Yao, M, Watanabe, M, Ohta, T, Tanaka, I, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2011-07-09 | | Release date: | 2012-02-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the enzyme CapF of Staphylococcus aureus reveals a unique architecture composed of two functional domains.
Biochem.J., 443, 2012
|
|
3VHR
 
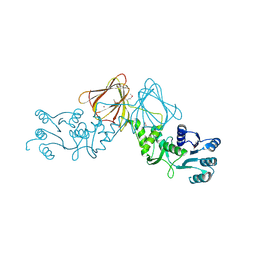 | | Crystal Structure of capsular polysaccharide assembling protein CapF from Staphylococcus aureus in space group C2221 | | Descriptor: | Capsular polysaccharide synthesis enzyme Cap5F, FORMIC ACID, ZINC ION | | Authors: | Miyafusa, T, Yoshikazu, T, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2011-09-01 | | Release date: | 2012-02-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the enzyme CapF of Staphylococcus aureus reveals a unique architecture composed of two functional domains.
Biochem.J., 443, 2012
|
|
5YD4
 
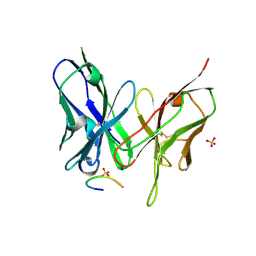 | |
5YD5
 
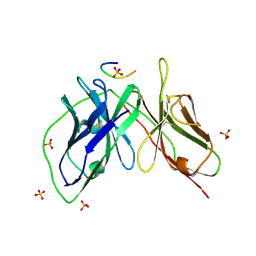 | |
