7JNI
 
 | | Crystal structure of the angiotensin II type 2 receptoror (AT2R) in complex with EMA401 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, FORMIC ACID, HEXANE-1,6-DIOL, ... | | Authors: | Cherezov, V, Shaye, H, Han, G.W. | | Deposit date: | 2020-08-04 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of the angiotensin II type 2 receptor AT 2 R is a novel therapeutic strategy for glioblastoma.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5GML
 
 | |
3Q2L
 
 | | Mouse E-cadherin EC1-2 V81D mutant | | Descriptor: | CALCIUM ION, Cadherin-1, PENTAETHYLENE GLYCOL | | Authors: | Harrison, O.J, Jin, X, Shapiro, L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Structure, 19, 2011
|
|
8BG3
 
 | |
8BG5
 
 | |
8BG4
 
 | |
8BG8
 
 | | SARS-CoV-2 S protein in complex with pT1696 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, pT1696 Fab heavy chain, ... | | Authors: | Hansen, G, Benecke, T, Vollmer, B, Gruenewald, K, Krey, T. | | Deposit date: | 2022-10-27 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Activity of broadly neutralizing antibodies against sarbecoviruses: a trade-off between SARS-CoV-2 variants and distant coronaviruses?
To be published
|
|
8BG6
 
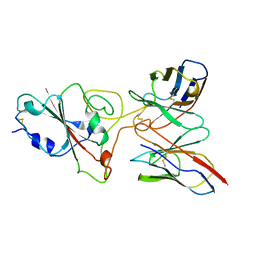 | | SARS-CoV-2 S protein in complex with pT1644 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, pT1644 Fab heavy chain, ... | | Authors: | Stroeh, L, Hansen, G, Vollmer, B, Krey, T, Benecke, T, Gruenewald, K. | | Deposit date: | 2022-10-27 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Activity of broadly neutralizing antibodies against sarbecoviruses: a trade-off between SARS-CoV-2 variants and distant coronaviruses?
To be published
|
|
4X3E
 
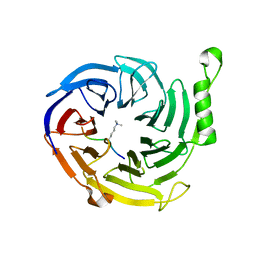 | |
3OOZ
 
 | | Bace1 in complex with the aminohydantoin Compound 102 | | Descriptor: | (5R)-2-amino-5-[4-(difluoromethoxy)phenyl]-5-[4-fluoro-3-(5-fluoropent-1-yn-1-yl)phenyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, Beta-secretase 1 | | Authors: | Olland, A.M. | | Deposit date: | 2010-08-31 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and Synthesis of Aminohydantoins as Potent and Selective Human beta-Secretase (BACE1) Inhibitors with Enhanced Brain Permeability
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3Q2V
 
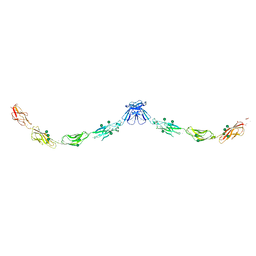 | | Crystal structure of mouse E-cadherin ectodomain | | Descriptor: | CALCIUM ION, Cadherin-1, MANGANESE (II) ION, ... | | Authors: | Jin, X, Harrison, O.J, Shapiro, L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Structure, 19, 2011
|
|
3Q2W
 
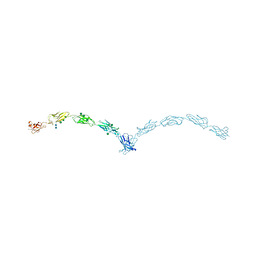 | | Crystal structure of mouse N-cadherin ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jin, X, Shapiro, L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Structure, 19, 2011
|
|
6SAK
 
 | |
2BRP
 
 | |
3Q2N
 
 | | Mouse E-cadherin EC1-2 L175D mutant | | Descriptor: | CALCIUM ION, Cadherin-1, TETRAETHYLENE GLYCOL | | Authors: | Harrison, O.J, Jin, X, Shapiro, L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Structure, 19, 2011
|
|
7A0N
 
 | | Structure of TSC1 NTD and linker domain | | Descriptor: | Uncharacterized protein,Uncharacterized protein | | Authors: | Fitzian, K, Kuemmel, D. | | Deposit date: | 2020-08-10 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | TSC1 binding to lysosomal PIPs is required for TSC complex translocation and mTORC1 regulation.
Mol.Cell, 81, 2021
|
|
7A0M
 
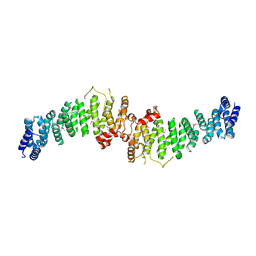 | | TSC1 N-terminal domain | | Descriptor: | SULFATE ION, TSC1 N-terminal domain | | Authors: | Zech, R, Kiontke, S, Kuemmel, D. | | Deposit date: | 2020-08-10 | | Release date: | 2021-05-26 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | TSC1 binding to lysosomal PIPs is required for TSC complex translocation and mTORC1 regulation.
Mol.Cell, 81, 2021
|
|
8P9X
 
 | | Vitamin D receptor complex with Xe4Me agonist ligand | | Descriptor: | (1~{R},3~{S},5~{Z})-5-[(2~{E})-2-[(1~{S},3~{a}~{S},7~{a}~{S})-1,7~{a}-dimethyl-1-(5-methyl-5-oxidanyl-hexa-1,3-diynyl)-2,3,3~{a},5,6,7-hexahydroinden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, ACETATE ION, Nuclear receptor coactivator 2, ... | | Authors: | Rochel, N. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A vitamin D-based strategy overcomes chemoresistance in prostate cancer.
Br.J.Pharmacol., 2024
|
|
5ZS3
 
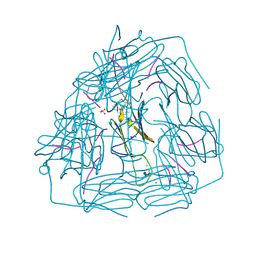 | | Small heat shock protein from M. marinum:Form-1 | | Descriptor: | CHLORIDE ION, GLY-ARG-LEU-LEU-PRO, Molecular chaperone (Small heat shock protein), ... | | Authors: | Bhandari, S, Suguna, K. | | Deposit date: | 2018-04-27 | | Release date: | 2019-01-30 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Dodecameric structure of a small heat shock protein from Mycobacterium marinum M.
Proteins, 87, 2019
|
|
5ZS6
 
 | |
5ZUL
 
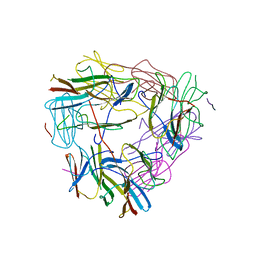 | |
3S7L
 
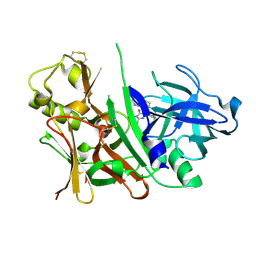 | |
8FEF
 
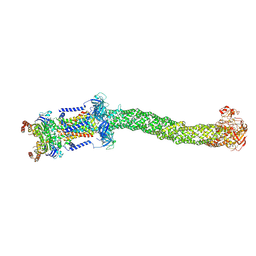 | | Structure of Mce1 transporter from Mycobacterium smegmatis (Map0) | | Descriptor: | ABC transporter, ATP-binding protein,Green fluorescent protein chimera, ABC-transporter integral membrane protein, ... | | Authors: | Chen, J, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structure of an endogenous mycobacterial MCE lipid transporter.
Nature, 620, 2023
|
|
8FED
 
 | | Structure of Mce1-LucB complex from Mycobacterium smegmatis (Map1) | | Descriptor: | ABC transporter, ATP-binding protein,Green fluorescent protein chimera, ABC-transporter integral membrane protein, ... | | Authors: | Chen, J, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structure of an endogenous mycobacterial MCE lipid transporter.
Nature, 620, 2023
|
|
8FEE
 
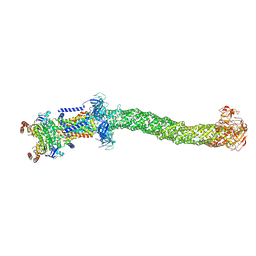 | | Structure of Mce1 transporter from Mycobacterium smegmatis in the absence of LucB (Map2) | | Descriptor: | ABC transporter, ATP-binding protein,Green fluorescent protein chimera, ABC-transporter integral membrane protein, ... | | Authors: | Chen, J, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of an endogenous mycobacterial MCE lipid transporter.
Nature, 620, 2023
|
|
