2CYH
 
 | | CYCLOPHILIN A COMPLEXED WITH DIPEPTIDE ALA-PRO | | Descriptor: | ALANINE, CYCLOPHILIN A, PROLINE | | Authors: | Zhao, Y, Ke, H. | | Deposit date: | 1996-02-27 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanistic implication of crystal structures of the cyclophilin-dipeptide complexes.
Biochemistry, 35, 1996
|
|
2BDM
 
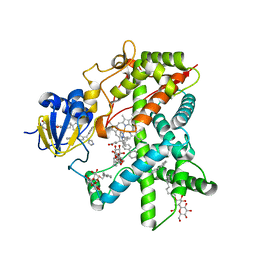 | | Structure of Cytochrome P450 2B4 with Bound Bifonazole | | Descriptor: | 1-[PHENYL-(4-PHENYLPHENYL)-METHYL]IMIDAZOLE, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, ... | | Authors: | Zhao, Y, White, M.A, Muralidhara, B.K, Sun, L, Halpert, J.R, Stout, C.D. | | Deposit date: | 2005-10-20 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of microsomal cytochrome P450 2B4 complexed with the antifungal drug bifonazole: insight into P450 conformational plasticity and membrane interaction.
J.Biol.Chem., 281, 2006
|
|
4Q4F
 
 | | Crystal structure of LIMP-2 (space group C2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-phosphono-beta-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Ren, J, Padilla-Parra, S, Fry, L.E, Stuart, D.I. | | Deposit date: | 2014-04-14 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lysosome sorting of beta-glucocerebrosidase by LIMP-2 is targeted by the mannose 6-phosphate receptor.
Nat Commun, 5, 2014
|
|
4Q4B
 
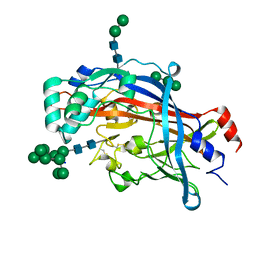 | | Crystal structure of LIMP-2 (space group C2221) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome membrane protein 2, ... | | Authors: | Zhao, Y, Ren, J, Padilla-Parra, S, Fry, L.E, Stuart, D.I. | | Deposit date: | 2014-04-14 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Lysosome sorting of beta-glucocerebrosidase by LIMP-2 is targeted by the mannose 6-phosphate receptor.
Nat Commun, 5, 2014
|
|
2MQS
 
 | |
2Q6N
 
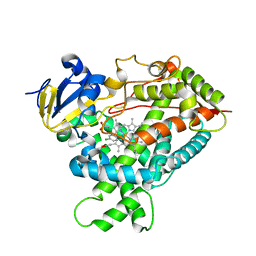 | | Structure of Cytochrome P450 2B4 with Bound 1-(4-cholorophenyl)imidazole | | Descriptor: | 1-(4-CHLOROPHENYL)-1H-IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhao, Y, Sun, L, Muralidhara, B.K, Kumar, S, White, M.A, Stout, C.D, Halpert, J.R. | | Deposit date: | 2007-06-05 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and thermodynamic consequences of 1-(4-chlorophenyl)imidazole binding to cytochrome P450 2B4.
Biochemistry, 46, 2007
|
|
4H4K
 
 | | Structure of the Cmr2-Cmr3 subcomplex of the Cmr RNA-silencing complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cmr subunit Cmr2, CRISPR system Cmr subunit Cmr3, ... | | Authors: | Shao, Y, Cocozaki, A.I, Ramia, N.F, Terns, R.M, Terns, M.P, Li, H. | | Deposit date: | 2012-09-17 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structure of the cmr2-cmr3 subcomplex of the cmr RNA silencing complex.
Structure, 21, 2013
|
|
4ILR
 
 | |
4ILM
 
 | | CRISPR RNA Processing endoribonuclease | | Descriptor: | CRISPR-associated endoribonuclease Cas6 2, RNA (5'-R(*GP*CP*UP*AP*AP*UP*CP*UP*AP*CP*UP*AP*UP*AP*GP*A)-3') | | Authors: | Shao, Y, Li, H. | | Deposit date: | 2012-12-31 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.068 Å) | | Cite: | Recognition and cleavage of a nonstructured CRISPR RNA by its processing endoribonuclease Cas6.
Structure, 21, 2013
|
|
4ILL
 
 | |
5ZG9
 
 | | Crystal structure of MoSub1-ssDNA complex in phosphate buffer | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*G)-3'), MoSub1, PHOSPHATE ION | | Authors: | Zhao, Y, Huang, J, Liu, H, Yi, L, Wang, S, Zhang, X, Liu, J. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The effect of phosphate ion on the ssDNA binding mode of MoSub1, a Sub1/PC4 homolog from rice blast fungus.
Proteins, 87, 2019
|
|
7X31
 
 | | solution structure of an anti-CRISPR protein | | Descriptor: | Anti-CRISPR protein (AcrIIC1) | | Authors: | Zhao, Y, Yang, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
6JP5
 
 | | Rabbit Cav1.1-Nifedipine Complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Huang, G, Wu, J, Yan, N. | | Deposit date: | 2019-03-25 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular Basis for Ligand Modulation of a Mammalian Voltage-Gated Ca2+Channel.
Cell, 177, 2019
|
|
6JP8
 
 | | Rabbit Cav1.1-Bay K8644 Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Huang, G, Wu, J, Yan, N. | | Deposit date: | 2019-03-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular Basis for Ligand Modulation of a Mammalian Voltage-Gated Ca2+Channel.
Cell, 177, 2019
|
|
6JPB
 
 | | Rabbit Cav1.1-Diltiazem Complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Huang, G, Wu, J, Yan, N. | | Deposit date: | 2019-03-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular Basis for Ligand Modulation of a Mammalian Voltage-Gated Ca2+Channel.
Cell, 177, 2019
|
|
6JPA
 
 | | Rabbit Cav1.1-Verapamil Complex | | Descriptor: | (2S)-2-(3,4-dimethoxyphenyl)-5-{[2-(3,4-dimethoxyphenyl)ethyl](methyl)amino}-2-(propan-2-yl)pentanenitrile, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Zhao, Y, Huang, G, Wu, J, Yan, N. | | Deposit date: | 2019-03-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular Basis for Ligand Modulation of a Mammalian Voltage-Gated Ca2+Channel.
Cell, 177, 2019
|
|
5WSZ
 
 | | Crystal structure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis | | Descriptor: | COPPER (II) ION, LpmO10A | | Authors: | Zhao, Y, Zhang, H, Yin, H. | | Deposit date: | 2016-12-08 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.565 Å) | | Cite: | Crystal structure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis
To Be Published
|
|
8JCN
 
 | |
8JCK
 
 | |
8JCM
 
 | |
8JCL
 
 | |
8JCO
 
 | |
8JCJ
 
 | |
8H3T
 
 | | The crystal structure of AlpH | | Descriptor: | AlpH, GLYCEROL | | Authors: | Zhao, Y, Li, M, Jiang, M, Pan, L.F. | | Deposit date: | 2022-10-09 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.866 Å) | | Cite: | O-methyltransferase-like enzyme catalyzed diazo installation in polyketide biosynthesis.
Nat Commun, 14, 2023
|
|
7D7L
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in complex with YM155 | | Descriptor: | 1-(2-methoxyethyl)-2-methyl-3-(pyrazin-2-ylmethyl)benzo[f]benzimidazol-3-ium-4,9-dione, CAFFEINE, GLYCEROL, ... | | Authors: | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2020-10-04 | | Release date: | 2021-04-21 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
