4K62
 
 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus A/Indonesia/5/2005 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Zhang, W, Shi, Y, Lu, X, Shu, Y, Qi, J, Gao, G.F. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level.
Science, 340, 2013
|
|
4K66
 
 | | Structure of an airborne transmissible avian influenza H5 hemagglutinin mutant from the influenza virus A/Indonesia/5/2005 complexed with avian receptor analog LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose | | Authors: | Zhang, W, Shi, Y, Lu, X, Shu, Y, Qi, J, Gao, G.F. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level.
Science, 340, 2013
|
|
4K63
 
 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus complexed with avian receptor analog LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Zhang, W, Shi, Y, Lu, X, Shu, Y, Qi, J, Gao, G.F. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level.
Science, 340, 2013
|
|
7VJT
 
 | | Crystal Structure of Mtb Pks13-TE in complex with inhibitor coumestan derivative 8 | | Descriptor: | 3,8-bis(oxidanyl)-7-(piperidin-1-ylmethyl)-[1]benzofuro[3,2-c]chromen-6-one, Polyketide synthase Pks13 (Termination polyketide synthase) | | Authors: | Zhang, W, Wang, S.S, Yu, L.F. | | Deposit date: | 2021-09-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-Based Optimization of Coumestan Derivatives as Polyketide Synthase 13-Thioesterase(Pks13-TE) Inhibitors with Improved hERG Profiles for Mycobacterium tuberculosis Treatment.
J.Med.Chem., 65, 2022
|
|
4K65
 
 | | Structure of an airborne transmissible avian influenza H5 hemagglutinin mutant from the influenza virus A/Indonesia/5/2005 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Zhang, W, Shi, Y, Lu, X, Shu, Y, Qi, J, Gao, G.F. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level.
Science, 340, 2013
|
|
4K64
 
 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus complexed with human receptor analog LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose | | Authors: | Zhang, W, Shi, Y, Lu, X, Shu, Y, Qi, J, Gao, G.F. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level.
Science, 340, 2013
|
|
4JTX
 
 | | Crystal structure of 2009 pandemic influenza virus hemagglutinin mutant D225E | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Zhang, W, Shi, Y, Qi, J, Gao, F, Li, Q, Fan, Z, Yan, J, Gao, G.F. | | Deposit date: | 2013-03-24 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Molecular basis of the receptor binding specificity switch of the hemagglutinins from both the 1918 and 2009 pandemic influenza A viruses by a D225G substitution
J.Virol., 87, 2013
|
|
4JTV
 
 | | Crystal structure of 2009 pandemic influenza virus hemagglutinin complexed with human receptor analogue LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Zhang, W, Shi, Y, Qi, J, Gao, F, Li, Q, Fan, Z, Yan, J, Gao, G.F. | | Deposit date: | 2013-03-24 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Molecular basis of the receptor binding specificity switch of the hemagglutinins from both the 1918 and 2009 pandemic influenza A viruses by a D225G substitution
J.Virol., 87, 2013
|
|
1U7R
 
 | |
6L16
 
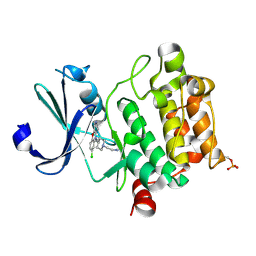 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-[4-[2-(7-chloranylpyrido[3,4-b][1,4]benzoxazin-5-yl)ethyl]piperidin-1-yl]ethanamine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L11
 
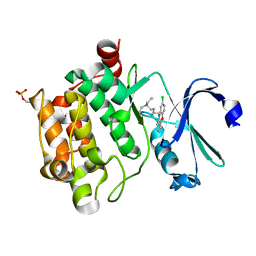 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 5-(2-chloranylphenoxazin-10-yl)-~{N},~{N}-diethyl-pentan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L17
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 7-chloranyl-5-[3-[(3~{S})-piperidin-3-yl]propyl]pyrido[3,4-b][1,4]benzoxazin-8-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L13
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-chloranyl-10-(2-piperidin-4-ylethyl)phenoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L14
 
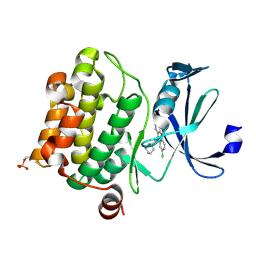 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-chloranyl-10-[3-[(3~{S})-piperidin-3-yl]propyl]phenoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
5FI4
 
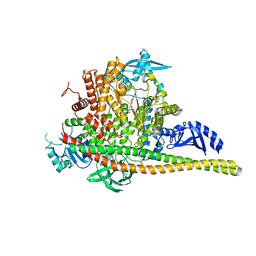 | | Discovery of imidazo[1,2-a]-pyridine inhibitors of pan-PI3 kinases that are efficacious in a mouse xenograft model | | Descriptor: | GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Elling, R.A, Knapp, M.S, Han, W, Daniel, L.M, Xy, Y, Burger, M.T, Ni, Z, Smith, A, Lan, J, Williams, T, Verhagen, J, Huh, K, Merritt, H, Chan, J, Kaufman, S, Voliva, C.F, Pecchi, S. | | Deposit date: | 2015-12-22 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of imidazo[1,2-a]-pyridine inhibitors of pan-PI3 kinases that are efficacious in a mouse xenograft model.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8DMF
 
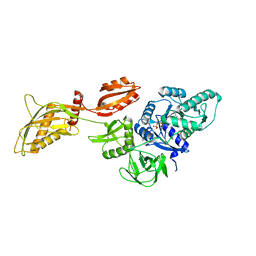 | | Cryo-EM structure of the ribosome-bound Bacteroides thetaiotaomicron EF-G2 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Tetracycline resistance protein TetQ | | Authors: | Wang, C, Han, W, Groisman, E.A, Liu, J. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-04 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Gut colonization by Bacteroides requires translation by an EF-G paralog lacking GTPase activity.
Embo J., 2022
|
|
5ZOF
 
 | | Crystal Structure of D181A/R192F hFen1 in complex with DNA | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*G)-3'), DNA (5'-D(*CP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*G)-3'), DNA (5'-D(*GP*CP*CP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
5ZOE
 
 | | Crystal Structure of D181A hFen1 in complex with DNA | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*G)-3'), DNA (5'-D(*CP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*G)-3'), DNA (5'-D(*GP*CP*CP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
5ZOD
 
 | | Crystal Structure of hFen1 in apo form | | Descriptor: | Flap endonuclease 1, MAGNESIUM ION, POTASSIUM ION | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
5ZOG
 
 | | Crystal Structure of R192F hFen1 in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*G)-3'), ... | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
1M9U
 
 | |
8SX6
 
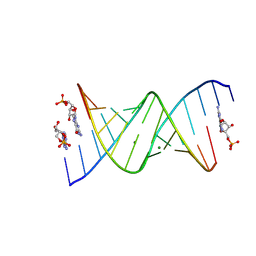 | | RNA duplex bound with GMP and AMP monomers | | Descriptor: | ADENOSINE MONOPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2023-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
8SWG
 
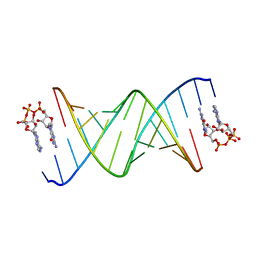 | | RNA duplex bound with GpppA dinucleotide ligand | | Descriptor: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G*(GA3))-3') | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
8SX5
 
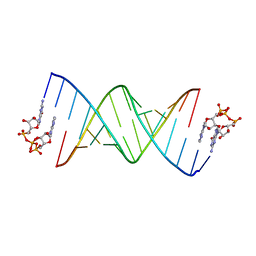 | | GpppA dinucleotide binding to RNA CU template | | Descriptor: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA (5'-R(*(TLN)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G*(GA3))-3') | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2023-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
8SWO
 
 | | GpppA dinucleotide ligand binding to RNA UC template | | Descriptor: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (5'-R(*(TLN)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G*(GA3))-3') | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2023-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
