2OL9
 
 | |
2OMP
 
 | |
2OLX
 
 | |
2ONA
 
 | |
2ONV
 
 | |
2OMM
 
 | |
2OMQ
 
 | |
3Q25
 
 | | Crystal structure of human alpha-synuclein (1-19) fused to maltose binding protein (MBP) | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein/alpha-synuclein chimeric protein, SULFATE ION, ... | | Authors: | Zhao, M, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2010-12-19 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of segments of alpha-synuclein fused to maltose-binding protein suggest intermediate states during amyloid formation
Protein Sci., 20, 2011
|
|
3Q28
 
 | | Cyrstal structure of human alpha-synuclein (58-79) fused to maltose binding protein (MBP) | | Descriptor: | Maltose-binding periplasmic protein/alpha-synuclein chimeric protein, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhao, M, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2010-12-19 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of segments of alpha-synuclein fused to maltose-binding protein suggest intermediate states during amyloid formation
Protein Sci., 20, 2011
|
|
3Q26
 
 | | Cyrstal structure of human alpha-synuclein (10-42) fused to maltose binding protein (MBP) | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein/alpha-synuclein chimeric protein, SULFATE ION, ... | | Authors: | Zhao, M, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2010-12-19 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structures of segments of alpha-synuclein fused to maltose-binding protein suggest intermediate states during amyloid formation
Protein Sci., 20, 2011
|
|
3Q29
 
 | | Cyrstal structure of human alpha-synuclein (1-19) fused to maltose binding protein (MBP) | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein/alpha-synuclein chimeric protein, SULFATE ION, ... | | Authors: | Zhao, M, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2010-12-19 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of segments of alpha-synuclein fused to maltose-binding protein suggest intermediate states during amyloid formation
Protein Sci., 20, 2011
|
|
3Q27
 
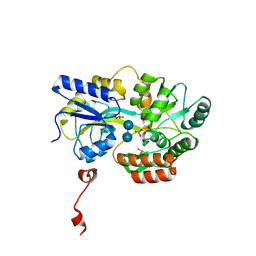 | | Cyrstal structure of human alpha-synuclein (32-57) fused to maltose binding protein (MBP) | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein/alpha-synuclein chimeric protein, SULFATE ION, ... | | Authors: | Zhao, M, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2010-12-19 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Structures of segments of alpha-synuclein fused to maltose-binding protein suggest intermediate states during amyloid formation
Protein Sci., 20, 2011
|
|
2ONW
 
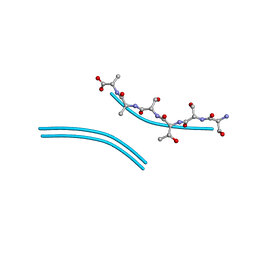 | |
3Q9G
 
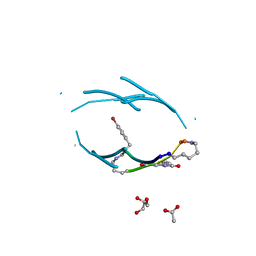 | | VQIVY segment from Alzheimer's tau displayed on 42-membered macrocycle scaffold | | Descriptor: | ACETIC ACID, Cyclic pseudo-peptide VQIV(4BF)(ORN)(HAO)KL(ORN), GLYCEROL | | Authors: | Liu, C, Sawaya, M.R, Eisenberg, D, Nowick, J.S, Cheng, P, Zheng, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic beta-Sheet Mimics.
J.Am.Chem.Soc., 133, 2011
|
|
3Q9J
 
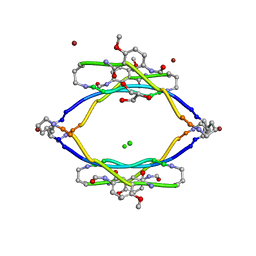 | | AIIFL segment derived from Alzheimer's Amyloid-Beta displayed on 42-membered macrocycle scaffold | | Descriptor: | CHLORIDE ION, Cyclic pseudo-peptide AIIFL(ORN)(HAO)YK(ORN), GLYCEROL, ... | | Authors: | Liu, C, Sawaya, M.R, Eisenberg, D, Nowick, J.S, Cheng, P, Zheng, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic beta-Sheet Mimics.
J.Am.Chem.Soc., 133, 2011
|
|
3NHD
 
 | | GYVLGS segment 127-132 from human prion with V129 | | Descriptor: | ACETIC ACID, Major prion protein | | Authors: | Apostol, M.I, Eisenberg, D. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystallographic studies of prion protein (PrP) segments suggest how structural changes encoded by polymorphism at residue 129 modulate susceptibility to human prion disease.
J.Biol.Chem., 285, 2010
|
|
1JRK
 
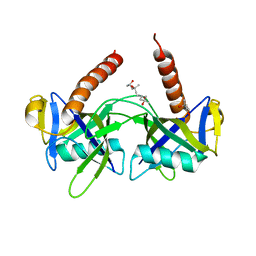 | | Crystal Structure of a Nudix Protein from Pyrobaculum aerophilum Reveals a Dimer with Intertwined Beta Sheets | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Nudix homolog | | Authors: | Wang, S, Mura, C, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2001-08-13 | | Release date: | 2002-04-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Nudix protein from Pyrobaculum aerophilum reveals a dimer with two intersubunit beta-sheets.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1F0X
 
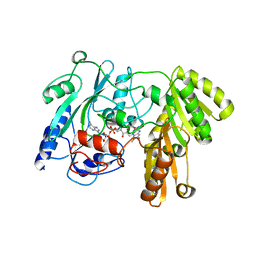 | | CRYSTAL STRUCTURE OF D-LACTATE DEHYDROGENASE, A PERIPHERAL MEMBRANE RESPIRATORY ENZYME. | | Descriptor: | D-LACTATE DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Dym, O, Pratt, E.A, Ho, C, Eisenberg, D. | | Deposit date: | 2000-05-17 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of D-lactate dehydrogenase, a peripheral membrane respiratory enzyme.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1F1H
 
 | |
3NVH
 
 | |
3NVE
 
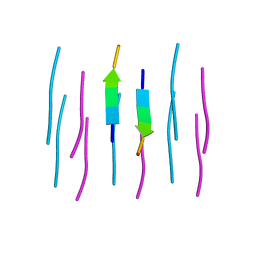 | |
3NVF
 
 | |
3NVG
 
 | |
1L9L
 
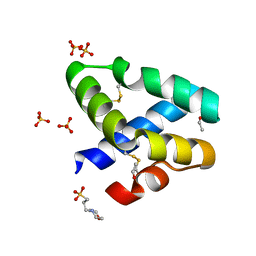 | | GRANULYSIN FROM HUMAN CYTOLYTIC T LYMPHOCYTES | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ETHANOL, Granulysin, ... | | Authors: | Anderson, D.H, Sawaya, M.R, Cascio, D, Ernst, W, Krensky, A, Modlin, R, Eisenberg, D. | | Deposit date: | 2002-03-25 | | Release date: | 2002-11-06 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Granulysin Crystal Structure and a Structure-Derived Lytic Mechanism
J.Mol.Biol., 325, 2002
|
|
3T4G
 
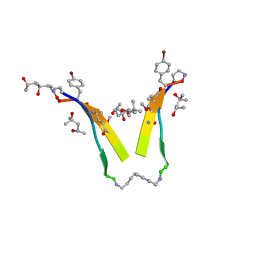 | | AIIGLMV segment from Alzheimer's Amyloid-Beta displayed on 54-membered macrocycle scaffold | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclic pseudo-peptide (ORN)AIIGLMV(ORN)KF(HAO)(4BF)K | | Authors: | Zhao, M, Liu, C, Cheng, P.N, Eisenberg, D, Nowick, J.S. | | Deposit date: | 2011-07-26 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Amyloid beta-sheet mimics that antagonize protein aggregation and reduce amyloid toxicity.
Nat Chem, 4, 2012
|
|
