6VII
 
 | |
6VIH
 
 | | The ligand-free structure of mouse RABL3 | | Descriptor: | Rab-like protein 3 | | Authors: | Su, L, Tomchick, D.R, Beutler, B. | | Deposit date: | 2020-01-13 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | Genetic and structural studies of RABL3 reveal an essential role in lymphoid development and function.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VIJ
 
 | | Crystal structure of mouse RABL3 in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Rab-like protein 3 | | Authors: | Su, L, Tomchick, D.R, Beutler, B. | | Deposit date: | 2020-01-13 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Genetic and structural studies of RABL3 reveal an essential role in lymphoid development and function.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VIK
 
 | |
1KQN
 
 | | Crystal structure of NMN/NaMN adenylyltransferase complexed with NAD | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, XENON | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Human Nicotinamide/Nicotonic Acid Mononucleotide Adenylyltransferase. Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2003
|
|
1KQO
 
 | | Crystal structure of NMN/NaMN adenylyltransferase complexed with deamido-NAD | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE, NICOTINIC ACID ADENINE DINUCLEOTIDE | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Hhuman of Nicotinamide/Nicotinic Acid Mononucleotide Adenylyltransferase.
Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2002
|
|
1KR2
 
 | | CRYSTAL STRUCTURE OF HUMAN NMN/NAMN ADENYLYL TRANSFERASE COMPLEXED WITH TIAZOFURIN ADENINE DINUCLEOTIDE (TAD) | | Descriptor: | BETA-METHYLENE-THIAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-08 | | Release date: | 2003-01-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Hhuman of Nicotinamide/Nicotinic Acid Mononucleotide Adenylyltransferase.
Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2002
|
|
1JPZ
 
 | | Crystal structure of a complex of the heme domain of P450BM-3 with N-Palmitoylglycine | | Descriptor: | BIFUNCTIONAL P-450:NADPH-P450 REDUCTASE, N-PALMITOYLGLYCINE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Haines, D.C, Tomchick, D.R, Machius, M, Peterson, J.A. | | Deposit date: | 2001-08-03 | | Release date: | 2001-11-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Pivotal role of water in the mechanism of P450BM-3.
Biochemistry, 40, 2001
|
|
1U3D
 
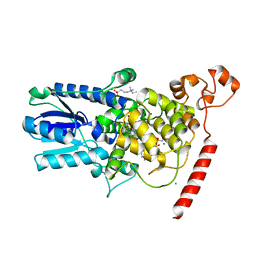 | | Crystal Structure of the PHR domain of Cryptochrome 1 from Arabidopsis thaliana with AMPPNP bound | | Descriptor: | CHLORIDE ION, Cryptochrome 1 apoprotein, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Brautigam, C.A, Smith, B.S, Ma, Z, Palnitkar, M, Tomchick, D.R, Machius, M, Deisenhofer, J. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1U3C
 
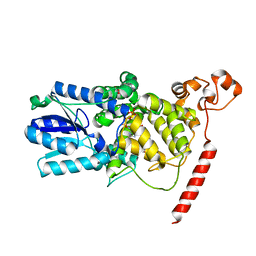 | | Crystal Structure of the PHR domain of Cryptochrome 1 from Arabidopsis thaliana | | Descriptor: | CHLORIDE ION, Cryptochrome 1 apoprotein, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Brautigam, C.A, Smith, B.S, Ma, Z, Palnitkar, M, Tomchick, D.R, Machius, M, Deisenhofer, J. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4QPM
 
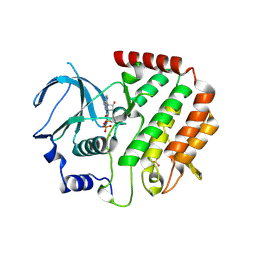 | | Structure of Bub1 kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Lin, Z.H, Jia, L.Y, Tomchick, D.R, Luo, X.L, Yu, H.T. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-22 | | Last modified: | 2014-12-24 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Substrate-Specific Activation of the Mitotic Kinase Bub1 through Intramolecular Autophosphorylation and Kinetochore Targeting.
Structure, 22, 2014
|
|
4R38
 
 | |
6MRW
 
 | | 14-meric ClyA pore complex | | Descriptor: | Hemolysin E, chromosomal | | Authors: | Peng, W, de Souza Santos, M, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | High-resolution cryo-EM structures of the E. coli hemolysin ClyA oligomers.
Plos One, 14, 2019
|
|
6MRT
 
 | | 12-meric ClyA pore complex | | Descriptor: | Hemolysin E, chromosomal | | Authors: | Peng, W, de Souza Santos, M, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | High-resolution cryo-EM structures of the E. coli hemolysin ClyA oligomers.
Plos One, 14, 2019
|
|
6MRU
 
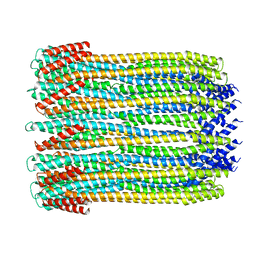 | | 13-meric ClyA pore complex | | Descriptor: | Hemolysin E, chromosomal | | Authors: | Peng, W, de Souza Santos, M, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution cryo-EM structures of the E. coli hemolysin ClyA oligomers.
Plos One, 14, 2019
|
|
4PJW
 
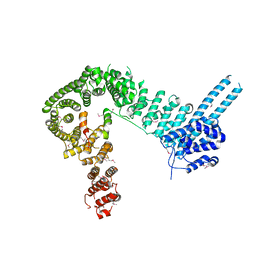 | | crystal structure of human Stromal Antigen 2 (SA2) in complex with Sister Chromatid Cohesion protein 1 (Scc1), with bound MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog | | Authors: | Hara, K, Chen, Z, Tomchick, D.R, Yu, H. | | Deposit date: | 2014-05-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4PJU
 
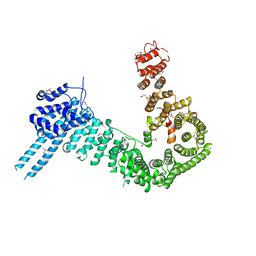 | | crystal structure of human Stromal Antigen 2 (SA2) in complex with Sister Chromatid Cohesion protein 1 (Scc1) | | Descriptor: | Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog | | Authors: | Hara, K, Chen, Z, Tomchick, D.R, Yu, H. | | Deposit date: | 2014-05-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4PK7
 
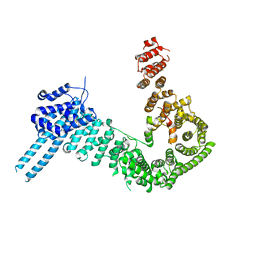 | | crystal structure of human Stromal Antigen 2 (SA2) in complex with Sister Chromatid Cohesion protein 1 (Scc1) with bound MES, native proteins | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog | | Authors: | Hara, K, Chen, Z, Tomchick, D.R, Yu, H. | | Deposit date: | 2014-05-13 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1NF3
 
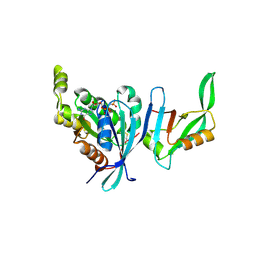 | | Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6 | | Descriptor: | G25K GTP-binding protein, placental isoform, MAGNESIUM ION, ... | | Authors: | Garrard, S.M, Capaldo, C.T, Gao, L, Rosen, M.K, Macara, I.G, Tomchick, D.R. | | Deposit date: | 2002-12-12 | | Release date: | 2003-03-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6
Embo J., 22, 2003
|
|
1OLS
 
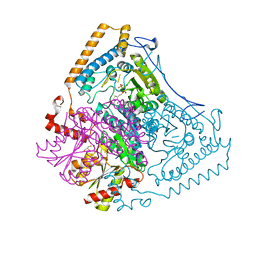 | | Roles of His291-alpha and His146-beta' in the reductive acylation reaction catalyzed by human branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, GLYCEROL, ... | | Authors: | Wynn, R.M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2003-08-12 | | Release date: | 2003-08-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Roles of His291-Alpha and His146-Beta' in the Reductive Acylation Reaction Catalyzed by Human Branched-Chain Alpha-Ketoacid Dehydrogenase: Refined Phosphorylation Loop Structure in the Active Site.
J.Biol.Chem., 278, 2003
|
|
2CMN
 
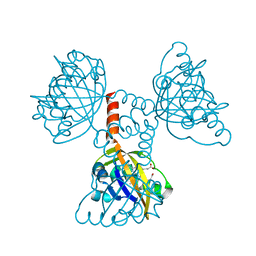 | | A Proximal Arginine Residue in the Switching Mechanism of the FixL Oxygen Sensor | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SENSOR PROTEIN FIXL | | Authors: | Gilles-Gonzalez, M.-A, Caceres, A.I, Silva Sousa, E.H, Tomchick, D.R, Brautigam, C.A, Gonzalez, C, Machius, M. | | Deposit date: | 2006-05-11 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Proximal Arginine R206 Participates in Switching of the Bradyrhizobium Japonicum Fixl Oxygen Sensor
J.Mol.Biol., 360, 2006
|
|
2BWQ
 
 | | Crystal Structure of the RIM2 C2A-domain at 1.4 angstrom Resolution | | Descriptor: | REGULATING SYNAPTIC MEMBRANE EXOCYTOSIS PROTEIN 2, SULFATE ION | | Authors: | Dai, H, Tomchick, D.R, Garcia, J, Sudhof, T.C, Machius, M, Rizo, J. | | Deposit date: | 2005-07-15 | | Release date: | 2005-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal Structure of the Rim2 C(2)A-Domain at 1.4 A Resolution.
Biochemistry, 44, 2005
|
|
2BNX
 
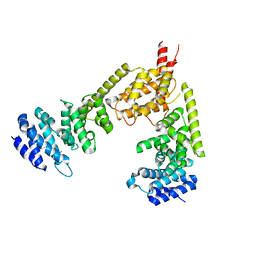 | | Crystal structure of the dimeric regulatory domain of mouse diaphaneous-related formin (DRF), mDia1 | | Descriptor: | CHLORIDE ION, DIAPHANOUS PROTEIN HOMOLOG 1 | | Authors: | Otomo, T, Otomo, C, Tomchick, D.R, Machius, M, Rosen, M.K. | | Deposit date: | 2005-04-05 | | Release date: | 2005-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Rho Gtpase-Mediated Activation of the Formin Mdia1
Mol.Cell, 18, 2005
|
|
2CJT
 
 | | Structural Basis for a Munc13-1 Homodimer - Munc13-1 - RIM Heterodimer Switch: C2-domains as Versatile Protein-Protein Interaction Modules | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, UNC-13 HOMOLOG A | | Authors: | Lu, J, Machius, M, Dulubova, I, Dai, H, Sudhof, T.C, Tomchick, D.R, Rizo, J. | | Deposit date: | 2006-04-06 | | Release date: | 2006-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Basis for a Munc13-1 Dimeric to Munc13-1/Rim Heterodimer Switch
Plos Biol., 4, 2006
|
|
2CJS
 
 | | Structural Basis for a Munc13-1 Homodimer - Munc13-1 - RIM Heterodimer Switch: C2-domains as Versatile Protein-Protein Interaction Modules | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, REGULATING SYNAPTIC MEMBRANE EXOCYTOSIS PROTEIN 2, ... | | Authors: | Lu, J, Machius, M, Dulubova, I, Dai, H, Sudhof, T.C, Tomchick, D.R, Rizo, J. | | Deposit date: | 2006-04-06 | | Release date: | 2006-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for a Munc13-1 Homodimer to Munc13-1/Rim Heterodimer Switch.
Plos Biol., 4, 2006
|
|
