5XJ5
 
 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the monoacylglycerol form | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, GLYCINE, Glycerol-3-phosphate acyltransferase, ... | | Authors: | Li, Z, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
4O9R
 
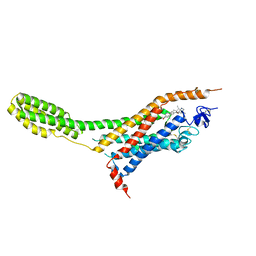 | | Human Smoothened Receptor structure in complex with cyclopamine | | Descriptor: | Cyclopamine, Smoothened homolog/Soluble cytochrome b562 chimeric protein | | Authors: | Wang, C, Weierstall, U, James, D, White, T.A, Wang, D, Liu, W, Spence, J.C.H, Doak, R.B, Nelson, G, Fromme, P, Fromme, R, Grotjohann, I, Kupitz, C, Zatsepin, N.A, Liu, H, Basu, S, Wacker, D, Han, G.W, Katritch, V, Boutet, S, Messerschmidt, M, Willams, G.J, Koglin, J.E, Seibert, M.M, Klinker, M, Gati, C, Shoeman, R.L, Barty, A, Chapman, H.N, Kirian, R.A, Beyerlein, K.R, Stevens, R.C, Li, D, Shah, S.T.A, Howe, N, Caffrey, M, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography.
Nat Commun, 5, 2014
|
|
8IMY
 
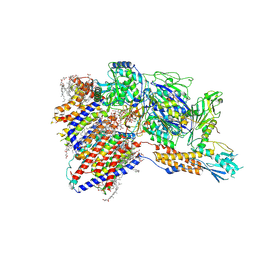 | | Cryo-EM structure of GPI-T (inactive mutant) with GPI and proULBP2, a proprotein substrate | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, T, Xu, Y, Qu, Q, Li, D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structures of liganded glycosylphosphatidylinositol transamidase illuminate GPI-AP biogenesis.
Nat Commun, 14, 2023
|
|
1I1H
 
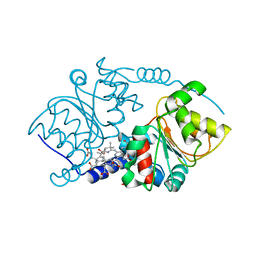 | | CRYSTAL STRUCTURE ANALYSIS OF PRECORRIN-8X METHYLMUTASE COMPLEX WITH HYDROGENOBYRINIC ACID | | Descriptor: | HYDROGENOBYRINIC ACID, PRECORRIN-8X METHYLMUTASE | | Authors: | Shipman, L.W, Li, D, Roessner, C.A, Scott, A.I, Sacchettini, J.C. | | Deposit date: | 2001-02-01 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of precorrin-8x methyl mutase.
Structure, 9, 2001
|
|
8K9R
 
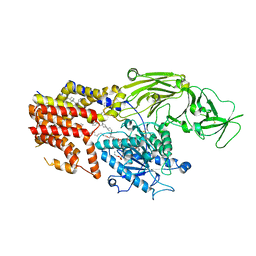 | | Cryo EM structure of the products-bound PGAP1(Bst1)-H443N from Chaetomium thermophilum | | Descriptor: | 2-amino-2-deoxy-alpha-D-glucopyranose, 2-azanylethyl [(2R,3S,4S,5S,6S)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hydrogen phosphate, 2-azanylethyl [(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl] hydrogen phosphate, ... | | Authors: | Li, T, Hong, J, Qu, Q, Li, D. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
8K9Q
 
 | | Cryo-EM structure of the GPI inositol-deacylase (PGAP1/Bst1) from Chaetomium thermophilum | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-3-hexadecanoyloxy-2-[(~{Z})-octadec-9-enoyl]oxy-propoxy]-oxidanyl-phosphoryl]oxy-propanoic acid, CHOLESTEROL HEMISUCCINATE, GPI inositol-deacylase,fused thermostable green fluorescent protein | | Authors: | Hong, J, Li, T, Qu, Q, Li, D. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
8K9T
 
 | | Cryo-EM structure of the products-bound PGAP1(Bst1)-S327A from Chaetonium thermophilum | | Descriptor: | 2-amino-2-deoxy-alpha-D-glucopyranose, 2-azanylethyl [(2R,3S,4S,5S,6S)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hydrogen phosphate, 2-azanylethyl [(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl] hydrogen phosphate, ... | | Authors: | Li, T, Hong, J, Qu, Q, Li, D. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
9J1R
 
 | | Structure of a triple-helix region of human Collagen type II from Trautec | | Descriptor: | SULFATE ION, Triple-helix region of human collagen type II | | Authors: | Fan, X, Chu, Y, Zhai, Y, Fu, S, Li, D, Cao, K, Feng, P, Wang, X, Le, H, Tang, D, Zhang, F, Qian, S. | | Deposit date: | 2024-08-05 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a triple-helix region of human Collagen type II from Trautec
To Be Published
|
|
6R2M
 
 | | Crystal structure of PssZ from Listeria monocytogenes | | Descriptor: | Glycoside transferase | | Authors: | Wu, H, Cheng, J, Qiao, S, Li, D, Ma, L. | | Deposit date: | 2019-03-18 | | Release date: | 2019-07-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | Crystal structure of the glycoside hydrolase PssZ from Listeria monocytogenes.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
8IMX
 
 | | Cryo-EM structure of GPI-T with a chimeric GPI-anchored protein | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, Y, Li, T, Qu, Q, Li, D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structures of liganded glycosylphosphatidylinositol transamidase illuminate GPI-AP biogenesis.
Nat Commun, 14, 2023
|
|
1GHQ
 
 | | CR2-C3D COMPLEX STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, COMPLEMENT C3, CR2/CD121/C3D/EPSTEIN-BARR VIRUS RECEPTOR, ... | | Authors: | Szakonyi, G, Guthridge, J.M, Li, D, Holers, V.M, Chen, X.S. | | Deposit date: | 2001-01-11 | | Release date: | 2001-06-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of complement receptor 2 in complex with its C3d ligand.
Science, 292, 2001
|
|
6J60
 
 | | hnRNP A1 reversible amyloid core GFGGNDNFG (residues 209-217) | | Descriptor: | 9-mer peptide (GFGGNDNFG) from Heterogeneous nuclear ribonucleoprotein A1 | | Authors: | Luo, F, Zhou, H, Gui, X, Li, D, Li, X, Liu, C. | | Deposit date: | 2019-01-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.96 Å) | | Cite: | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
6KJ4
 
 | | 120kV MicroED structure of FUS (37-42) SYSGYS solved from single crystal at 0.65 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.65 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
5X41
 
 | | 3.5A resolution structure of a cobalt energy-coupling factor transporter using LCP method-CbiMQO | | Descriptor: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | Authors: | Bao, Z, Qi, X, Zhao, W, Li, D, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
6KJ3
 
 | | 120kV MicroED structure of FUS (37-42) SYSGYS solved from merged datasets at 0.60 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.6 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
6KJ2
 
 | | 200kV MicroED structure of FUS (37-42) SYSGYS solved from single crystal at 0.67 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.67 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
8IK7
 
 | |
8IKS
 
 | |
8IKB
 
 | |
8IKP
 
 | |
8IHV
 
 | | Cryo-EM structure of an amyloid fibril formed by ALS-causing SOD1 mutation H46R | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Wang, L.Q, Ma, Y.Y, Zhang, M.Y, Yuan, H.Y, Li, X.N, Zhao, K, Chen, J, Li, D, Wang, Z.Z, Le, W.D, Liu, C, Liang, Y. | | Deposit date: | 2023-02-23 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Amyloid fibril structures and ferroptosis activation induced by ALS-causing SOD1 mutations.
Sci Adv, 2024
|
|
8IHU
 
 | | Cryo-EM structure of an amyloid fibril formed by ALS-causing SOD1 mutation G85R | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Wang, L.Q, Ma, Y.Y, Zhang, M.Y, Yuan, H.Y, Li, X.N, Zhao, K, Chen, J, Li, D, Wang, Z.Z, Le, W.D, Liu, C, Liang, Y. | | Deposit date: | 2023-02-23 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Amyloid fibril structures and ferroptosis activation induced by ALS-causing SOD1 mutations.
Sci Adv, 2024
|
|
8Z8Z
 
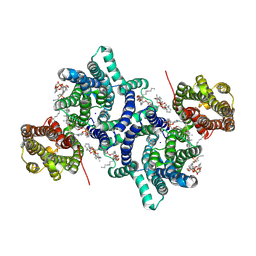 | | Cryo-EM structure of LYCHOS | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Xiong, Q, Zhu, Z, Li, T, Zhou, Z, Chao, Y, Qu, Q, Li, D. | | Deposit date: | 2024-04-22 | | Release date: | 2025-01-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Molecular architecture of human LYCHOS involved in lysosomal cholesterol signaling.
Nat.Struct.Mol.Biol., 32, 2025
|
|
7DWV
 
 | | Cryo-EM structure of amyloid fibril formed by familial prion disease-related mutation E196K | | Descriptor: | Major prion protein | | Authors: | Wang, L.Q, Zhao, K, Yuan, H.Y, Li, X.N, Dang, H.B, Ma, Y.Y, Wang, Q, Wang, C, Sun, Y.P, Chen, J, Li, D, Zhang, D.L, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2021-01-18 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Genetic prion disease-related mutation E196K displays a novel amyloid fibril structure revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7D2Z
 
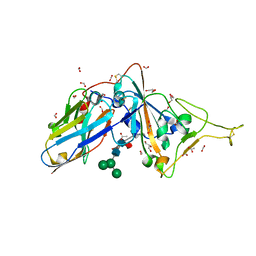 | | Structure of sybody SR31 in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Li, T, Cai, H, Yao, H, Qin, W, Li, D. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A high-affinity RBD-targeting nanobody improves fusion partner's potency against SARS-CoV-2.
Plos Pathog., 17, 2021
|
|
